The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin
- PMID: 20595233
- PMCID: PMC2895200
- DOI: 10.1101/gad.1906410
The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin
Abstract
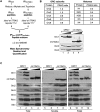
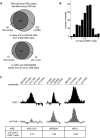
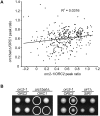
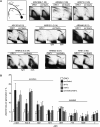
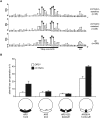
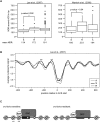
The origin recognition complex (ORC) binds to the specific positions on chromosomes that serve as DNA replication origins. Although ORC is conserved from yeast to humans, the DNA sequence elements that specify ORC binding are not. In particular, metazoan ORC shows no obvious DNA sequence specificity, whereas yeast ORC binds to a specific DNA sequence within all yeast origins. Thus, whereas chromatin must play an important role in metazoan ORC's ability to recognize origins, it is unclear whether chromatin plays a role in yeast ORC's recognition of origins. This study focused on the role of the conserved N-terminal bromo-adjacent homology domain of yeast Orc1 (Orc1BAH). Recent studies indicate that BAH domains are chromatin-binding modules. We show that the Orc1BAH domain was necessary for ORC's stable association with yeast chromosomes, and was physiologically relevant to DNA replication in vivo. This replication role was separable from the Orc1BAH domain's previously defined role in transcriptional silencing. Genome-wide analyses of ORC binding in ORC1 and orc1bahDelta cells revealed that the Orc1BAH domain contributed to ORC's association with most yeast origins, including a class of origins highly dependent on the Orc1BAH domain for ORC association (orc1bahDelta-sensitive origins). Orc1bahDelta-sensitive origins required the Orc1BAH domain for normal activity on chromosomes and plasmids, and were associated with a distinct local nucleosome structure. These data provide molecular insights into how the Orc1BAH domain contributes to ORC's selection of replication origins, as well as new tools for examining conserved mechanisms governing ORC's selection of origins within eukaryotic chromosomes.
Figures

Similar articles
-
The budding yeast Fkh1 Forkhead associated (FHA) domain promotes a G1-chromatin state and the activity of chromosomal DNA replication origins.PLoS Genet. 2024 Aug 5;20(8):e1011366. doi: 10.1371/journal.pgen.1011366. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39102423 Free PMC article.
-
The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo.EMBO J. 2006 Nov 15;25(22):5372-82. doi: 10.1038/sj.emboj.7601396. Epub 2006 Oct 26. EMBO J. 2006. PMID: 17066079 Free PMC article.
-
Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues.Sci Rep. 2015 Oct 12;5:14929. doi: 10.1038/srep14929. Sci Rep. 2015. PMID: 26456755 Free PMC article.
-
Compact Origins and Where to Find Them: ORC's Guide to Genome-Wide Licensing.Bioessays. 2025 Jul;47(7):e70018. doi: 10.1002/bies.70018. Epub 2025 May 19. Bioessays. 2025. PMID: 40384310 Free PMC article. Review.
-
The many faces of the origin recognition complex.Curr Opin Cell Biol. 2007 Jun;19(3):337-43. doi: 10.1016/j.ceb.2007.04.007. Epub 2007 Apr 26. Curr Opin Cell Biol. 2007. PMID: 17466500 Review.
Cited by
-
DNA sequence templates adjacent nucleosome and ORC sites at gene amplification origins in Drosophila.Nucleic Acids Res. 2015 Oct 15;43(18):8746-61. doi: 10.1093/nar/gkv766. Epub 2015 Jul 30. Nucleic Acids Res. 2015. PMID: 26227968 Free PMC article.
-
Getting there: understanding the chromosomal recruitment of the AAA+ ATPase Pch2/TRIP13 during meiosis.Curr Genet. 2021 Aug;67(4):553-565. doi: 10.1007/s00294-021-01166-3. Epub 2021 Mar 12. Curr Genet. 2021. PMID: 33712914 Free PMC article. Review.
-
Molecular determinants of origin discrimination by Orc1 initiators in archaea.Nucleic Acids Res. 2011 May;39(9):3621-31. doi: 10.1093/nar/gkq1308. Epub 2011 Jan 11. Nucleic Acids Res. 2011. PMID: 21227921 Free PMC article.
-
Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex.Mol Biol Cell. 2012 Jan;23(1):200-12. doi: 10.1091/mbc.E11-05-0409. Epub 2011 Nov 2. Mol Biol Cell. 2012. PMID: 22049023 Free PMC article.
-
The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses.Biochim Biophys Acta. 2012 Jul;1819(7):794-801. doi: 10.1016/j.bbagrm.2012.01.017. Epub 2012 Feb 8. Biochim Biophys Acta. 2012. PMID: 22342530 Free PMC article. Review.
References
-
- Aladjem MI 2007. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8: 588–600 - PubMed
-
- Bell SP 2002. The origin recognition complex: From simple origins to complex functions. Genes Dev 16: 659–672 - PubMed
-
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 - PubMed
-
- Bell SP, Kobayashi R, Stillman B 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262: 1844–1849 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
