A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana
- PMID: 20595237
- PMCID: PMC2921208
- DOI: 10.1093/jxb/erq188
A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana
Abstract
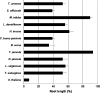
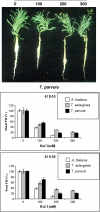
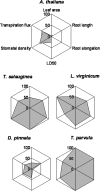
Salinity is an abiotic stress that limits both yield and the expansion of agricultural crops to new areas. In the last 20 years our basic understanding of the mechanisms underlying plant tolerance and adaptation to saline environments has greatly improved owing to active development of advanced tools in molecular, genomics, and bioinformatics analyses. However, the full potential of investigative power has not been fully exploited, because the use of halophytes as model systems in plant salt tolerance research is largely neglected. The recent introduction of halophytic Arabidopsis-Relative Model Species (ARMS) has begun to compare and relate several unique genetic resources to the well-developed Arabidopsis model. In a search for candidates to begin to understand, through genetic analyses, the biological bases of salt tolerance, 11 wild relatives of Arabidopsis thaliana were compared: Barbarea verna, Capsella bursa-pastoris, Hirschfeldia incana, Lepidium densiflorum, Malcolmia triloba, Lepidium virginicum, Descurainia pinnata, Sisymbrium officinale, Thellungiella parvula, Thellungiella salsuginea (previously T. halophila), and Thlaspi arvense. Among these species, highly salt-tolerant (L. densiflorum and L. virginicum) and moderately salt-tolerant (M. triloba and H. incana) species were identified. Only T. parvula revealed a true halophytic habitus, comparable to the better studied Thellungiella salsuginea. Major differences in growth, water transport properties, and ion accumulation are observed and discussed to describe the distinctive traits and physiological responses that can now be studied genetically in salt stress research.
Figures






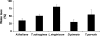

Similar articles
-
Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila.J Proteome Res. 2010 May 7;9(5):2584-99. doi: 10.1021/pr100034f. J Proteome Res. 2010. PMID: 20377188
-
Salt tolerance mechanisms in three Irano-Turanian Brassicaceae halophytes relatives of Arabidopsis thaliana.J Plant Res. 2018 Nov;131(6):1029-1046. doi: 10.1007/s10265-018-1053-6. Epub 2018 Jul 2. J Plant Res. 2018. PMID: 29967980
-
Differences in the Temporal Kinetics of the Metabolic Responses to Salinity Between the Salt-Tolerant Thellungiella salsuginea and the Salt-Sensitive Arabidopsis thaliana Reveal New Insights in Salt Tolerance Mechanisms.Int J Mol Sci. 2025 May 27;26(11):5141. doi: 10.3390/ijms26115141. Int J Mol Sci. 2025. PMID: 40507952 Free PMC article.
-
Halophytism: What Have We Learnt From Arabidopsis thaliana Relative Model Systems?Plant Physiol. 2018 Nov;178(3):972-988. doi: 10.1104/pp.18.00863. Epub 2018 Sep 20. Plant Physiol. 2018. PMID: 30237204 Free PMC article. Review.
-
Halophytes as new model plant species for salt tolerance strategies.Front Plant Sci. 2023 May 11;14:1137211. doi: 10.3389/fpls.2023.1137211. eCollection 2023. Front Plant Sci. 2023. PMID: 37251767 Free PMC article. Review.
Cited by
-
The High-Affinity Potassium Transporter EpHKT1;2 From the Extremophile Eutrema parvula Mediates Salt Tolerance.Front Plant Sci. 2018 Jul 30;9:1108. doi: 10.3389/fpls.2018.01108. eCollection 2018. Front Plant Sci. 2018. PMID: 30105045 Free PMC article.
-
Responses to Salinity in Four Plantago Species from Tunisia.Plants (Basel). 2021 Jul 7;10(7):1392. doi: 10.3390/plants10071392. Plants (Basel). 2021. PMID: 34371595 Free PMC article.
-
Salt Induces Features of a Dormancy-Like State in Seeds of Eutrema (Thellungiella) salsugineum, a Halophytic Relative of Arabidopsis.Front Plant Sci. 2016 Aug 3;7:1071. doi: 10.3389/fpls.2016.01071. eCollection 2016. Front Plant Sci. 2016. PMID: 27536302 Free PMC article.
-
Landscape of gene transposition-duplication within the Brassicaceae family.DNA Res. 2019 Feb 1;26(1):21-36. doi: 10.1093/dnares/dsy035. DNA Res. 2019. PMID: 30380026 Free PMC article.
-
The genome of the extremophile crucifer Thellungiella parvula.Nat Genet. 2011 Aug 7;43(9):913-8. doi: 10.1038/ng.889. Nat Genet. 2011. PMID: 21822265 Free PMC article.
References
-
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42.
-
- Al-Shehbaz IA, O'Kane SL. Placement of Arabidopsis parvula in Thellungiella (Brassicaceae) Novon. 1995;5:310.
-
- Aksoy A, Hale WHG, Dixon JM. Capsella bursa-pastoris (L.) Medic. as a biomonitor of heavy metal pollution. Science Total Environment. 1999;226:177–186. - PubMed

