An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b
- PMID: 20595614
- PMCID: PMC2935300
- DOI: 10.1126/science.1188989
An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b
Abstract
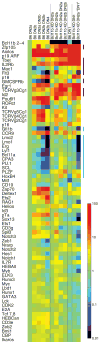
The identities of the regulators that mediate commitment of hematopoietic precursors to the T lymphocyte lineage have been unknown. The last stage of T lineage commitment in vivo involves mechanisms to suppress natural killer cell potential, to suppress myeloid and dendritic cell potential, and to silence the stem cell or progenitor cell regulatory functions that initially provide T cell receptor-independent self-renewal capability. The zinc finger transcription factor Bcl11b is T cell-specific in expression among hematopoietic cell types and is first expressed in precursors immediately before T lineage commitment. We found that Bcl11b is necessary for T lineage commitment in mice and is specifically required both to repress natural killer cell-associated genes and to down-regulate a battery of stem cell or progenitor cell genes at the pivotal stage of commitment.
Figures


Comment in
-
Immunology. A guardian of T cell fate.Science. 2010 Jul 2;329(5987):44-5. doi: 10.1126/science.1191664. Science. 2010. PMID: 20595605 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

