Influenza A subtyping: seasonal H1N1, H3N2, and the appearance of novel H1N1
- PMID: 20595627
- PMCID: PMC2928431
- DOI: 10.2353/jmoldx.2010.090225
Influenza A subtyping: seasonal H1N1, H3N2, and the appearance of novel H1N1
Abstract
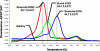
Influenza virus subtyping has emerged as a critical tool in the diagnosis of influenza. Antiviral resistance is present in the majority of seasonal H1N1 influenza A infections, with association of viral strain type and antiviral resistance. Influenza A virus subtypes can be reliably distinguished by examining conserved sequences in the matrix protein gene. We describe our experience with an assay for influenza A subtyping based on matrix gene sequences. Viral RNA was prepared from nasopharyngeal swab samples, and real-time RT-PCR detection of influenza A and B was performed using a laboratory developed analyte-specific reagent-based assay that targets a conserved region of the influenza A matrix protein gene. FluA-positive samples were analyzed using a second RT-PCR assay targeting the matrix protein gene to distinguish seasonal influenza subtypes based on differential melting of fluorescence resonance energy transfer probes. The novel H1N1 influenza strain responsible for the 2009 pandemic showed a melting profile distinct from that of seasonal H1N1 or H3N2 and compatible with the predicted melting temperature based on the published novel H1N1 matrix gene sequence. Validation by comparison with the Centers for Disease Control and Prevention real-time RT-PCR for swine influenza A (novel H1N1) test showed this assay to be both rapid and reliable (>99% sensitive and specific) in the identification of the novel H1N1 influenza A virus strain.
Figures
Similar articles
-
Rapid multiplex reverse transcription-PCR typing of influenza A and B virus, and subtyping of influenza A virus into H1, 2, 3, 5, 7, 9, N1 (human), N1 (animal), N2, and N7, including typing of novel swine origin influenza A (H1N1) virus, during the 2009 outbreak in Milwaukee, Wisconsin.J Clin Microbiol. 2009 Sep;47(9):2772-8. doi: 10.1128/JCM.00998-09. Epub 2009 Jul 29. J Clin Microbiol. 2009. PMID: 19641063 Free PMC article.
-
Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays.Clin Chem. 2009 Aug;55(8):1555-8. doi: 10.1373/clinchem.2009.130229. Epub 2009 May 13. Clin Chem. 2009. PMID: 19439731 Free PMC article.
-
One-step real-time reverse transcription-PCR assays for detecting and subtyping pandemic influenza A/H1N1 2009, seasonal influenza A/H1N1, and seasonal influenza A/H3N2 viruses.J Virol Methods. 2011 Jan;171(1):156-62. doi: 10.1016/j.jviromet.2010.10.018. Epub 2010 Oct 26. J Virol Methods. 2011. PMID: 21029748 Free PMC article.
-
Subtype identification of the novel A H1N1 and other human influenza A viruses using an oligonucleotide microarray.Arch Virol. 2010;155(1):55-61. doi: 10.1007/s00705-009-0545-z. Epub 2009 Dec 10. Arch Virol. 2010. PMID: 19998047
-
Rapid differential detection of subtype H1 and H3 swine influenza viruses using a TaqMan-MGB-based duplex one-step real-time RT-PCR assay.Arch Virol. 2021 Aug;166(8):2217-2224. doi: 10.1007/s00705-021-05127-6. Epub 2021 Jun 6. Arch Virol. 2021. PMID: 34091783
Cited by
-
Laboratories and Pandemic Preparedness: A Framework for Collaboration and Oversight.J Mol Diagn. 2020 Jul;22(7):841-843. doi: 10.1016/j.jmoldx.2020.05.002. Epub 2020 May 16. J Mol Diagn. 2020. PMID: 32425699 Free PMC article. No abstract available.
-
Analytical performance determination and clinical validation of the novel Roche RealTime Ready Influenza A/H1N1 Detection Set.J Clin Microbiol. 2010 Sep;48(9):3088-94. doi: 10.1128/JCM.00785-10. Epub 2010 Jul 7. J Clin Microbiol. 2010. PMID: 20610677 Free PMC article.
-
Subtyping influenza A virus with monoclonal antibodies and an indirect immunofluorescence assay.J Clin Microbiol. 2012 Feb;50(2):396-400. doi: 10.1128/JCM.01237-11. Epub 2011 Nov 9. J Clin Microbiol. 2012. PMID: 22075584 Free PMC article.
-
Therapeutic p28 peptide targets essential H1N1 influenza virus proteins: insights from docking and molecular dynamics simulations.Mol Divers. 2021 Aug;25(3):1929-1943. doi: 10.1007/s11030-021-10193-8. Epub 2021 Feb 11. Mol Divers. 2021. PMID: 33575983 Free PMC article.
-
Analysis of the Genetic Diversity Associated With the Drug Resistance and Pathogenicity of Influenza A Virus Isolated in Bangladesh From 2002 to 2019.Front Microbiol. 2021 Sep 17;12:735305. doi: 10.3389/fmicb.2021.735305. eCollection 2021. Front Microbiol. 2021. PMID: 34603265 Free PMC article. Review.
References
-
- Stone B, Burrows J, Schepetiuk S, Higgins G, Hampson A, Shaw R, Kok T. Rapid detection and simultaneous subtype differentiation of influenza A viruses by real time PCR. J Virol Methods. 2004;117:103–112. - PubMed
-
- Update: influenza activity—United States, September 28–November 29, 2008. MMWR Morb Mortal Wkly Rep. 2008;57:1329–1332. - PubMed
-
- CDC Human Influenza Virus Real-time RT-PCR Detection and Characterization Panel (rRT-PCR Flu Panel) Centers for Disease Control and Prevention; Atlanta, GA: 2008.
-
- CDC Protocol of Realtime RTPCR for Influenza A(H1N1) Centers for Disease Control and Prevention; Atlanta, GA: 2009.
-
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

