Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis
- PMID: 20595680
- PMCID: PMC3013519
- DOI: 10.1681/ASN.2009121244
Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis
Abstract
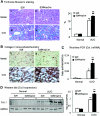
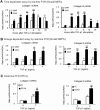
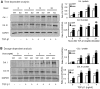
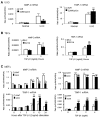
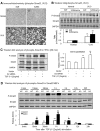
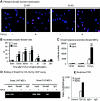
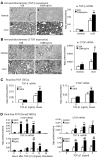
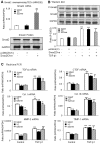
Smad2 and Smad3 interact and mediate TGF-beta signaling. Although Smad3 promotes fibrosis, the role of Smad2 in fibrogenesis is largely unknown. In this study, conditional deletion of Smad2 from the kidney tubular epithelial cells markedly enhanced fibrosis in response to unilateral ureteral obstruction. In vitro, Smad2 knockdown in tubular epithelial cells increased expression of collagen I, collagen III, and TIMP-1 and decreased expression of the matrix-degrading enzyme MMP-2 in response to TGF-beta1 compared with similarly treated wild-type cells. We obtained similar results in Smad2-knockout fibroblasts. Mechanistically, Smad2 deletion promoted fibrosis through enhanced TGF-beta/Smad3 signaling, evidenced by greater Smad3 phosphorylation, nuclear translocation, promoter activity, and binding of Smad3 to a collagen promoter (COL1A2). Moreover, deletion of Smad2 increased autoinduction of TGF-beta1. Conversely, overexpression of Smad2 attenuated TGF-beta1-induced Smad3 phosphorylation and collagen I matrix expression in tubular epithelial cells. In conclusion, in contrast to Smad3, Smad2 protects against TGF-beta-mediated fibrosis by counteracting TGF-beta/Smad3 signaling.
Figures


Similar articles
-
Smad2 protects against TGF-β1/Smad3-mediated collagen synthesis in human hepatic stellate cells during hepatic fibrosis.Mol Cell Biochem. 2015 Feb;400(1-2):17-28. doi: 10.1007/s11010-014-2258-1. Epub 2014 Oct 29. Mol Cell Biochem. 2015. PMID: 25351340
-
Angiotensin II induces connective tissue growth factor and collagen I expression via transforming growth factor-beta-dependent and -independent Smad pathways: the role of Smad3.Hypertension. 2009 Oct;54(4):877-84. doi: 10.1161/HYPERTENSIONAHA.109.136531. Epub 2009 Aug 10. Hypertension. 2009. PMID: 19667256
-
Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction.J Clin Invest. 2003 Nov;112(10):1486-94. doi: 10.1172/JCI19270. J Clin Invest. 2003. PMID: 14617750 Free PMC article.
-
LncRNA GAS5 protects against TGF-β-induced renal fibrosis via the Smad3/miRNA-142-5p axis.Am J Physiol Renal Physiol. 2021 Oct 1;321(4):F517-F526. doi: 10.1152/ajprenal.00085.2021. Epub 2021 Sep 6. Am J Physiol Renal Physiol. 2021. PMID: 34486400
-
TGF-β-Smad3 signaling in emphysema and pulmonary fibrosis: an epigenetic aberration of normal development?Am J Physiol Lung Cell Mol Physiol. 2013 Jan 15;304(2):L83-5. doi: 10.1152/ajplung.00258.2012. Epub 2012 Nov 16. Am J Physiol Lung Cell Mol Physiol. 2013. PMID: 23161884 Free PMC article. Review.
Cited by
-
Differential expression and therapeutic efficacy of microRNA-346 in diabetic nephropathy mice.Exp Ther Med. 2015 Jul;10(1):106-112. doi: 10.3892/etm.2015.2468. Epub 2015 Apr 30. Exp Ther Med. 2015. PMID: 26170919 Free PMC article.
-
Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity.Sci Rep. 2016 Oct 5;6:34790. doi: 10.1038/srep34790. Sci Rep. 2016. PMID: 27703224 Free PMC article.
-
RGMb protects against acute kidney injury by inhibiting tubular cell necroptosis via an MLKL-dependent mechanism.Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):E1475-E1484. doi: 10.1073/pnas.1716959115. Epub 2018 Jan 30. Proc Natl Acad Sci U S A. 2018. PMID: 29382757 Free PMC article.
-
L-Endoglin overexpression increases renal fibrosis after unilateral ureteral obstruction.PLoS One. 2014 Oct 14;9(10):e110365. doi: 10.1371/journal.pone.0110365. eCollection 2014. PLoS One. 2014. PMID: 25313562 Free PMC article.
-
Adipocyte-derived exosomes from obstructive sleep apnoea rats aggravate MASLD by TCONS_00039830/miR-455-3p/Smad2 axis.Commun Biol. 2024 Apr 23;7(1):492. doi: 10.1038/s42003-024-06171-z. Commun Biol. 2024. PMID: 38654054 Free PMC article.
References
-
- Schnaper HW, Hayashida T, Poncelet AC: It's a Smad world: Regulation of TGF-beta signaling in the kidney. J Am Soc Nephrol 13: 1126–1128, 2002 - PubMed
-
- Liu Y: Renal fibrosis: New insights into the pathogenesis and therapeutics. Kidney Int 69: 213–217, 2006 - PubMed
-
- Bottinger EP, Bitzer M: TGF-beta signaling in renal disease. J Am Soc Nephrol 13: 2600–2610, 2002 - PubMed
-
- Wang W, Koka V, Lan HY: Transforming growth factor-beta and Smad signalling in kidney diseases. Nephrology (Carlton) 10: 48–56, 2005 - PubMed
-
- Derynck R, Zhang YE: Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425: 577–584, 2003 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

