Genome-wide association study in alopecia areata implicates both innate and adaptive immunity
- PMID: 20596022
- PMCID: PMC2921172
- DOI: 10.1038/nature09114
Genome-wide association study in alopecia areata implicates both innate and adaptive immunity
Abstract
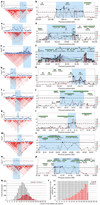
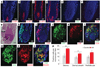
Alopecia areata (AA) is among the most highly prevalent human autoimmune diseases, leading to disfiguring hair loss due to the collapse of immune privilege of the hair follicle and subsequent autoimmune attack. The genetic basis of AA is largely unknown. We undertook a genome-wide association study (GWAS) in a sample of 1,054 cases and 3,278 controls and identified 139 single nucleotide polymorphisms that are significantly associated with AA (P <or= 5 x 10(-7)). Here we show an association with genomic regions containing several genes controlling the activation and proliferation of regulatory T cells (T(reg) cells), cytotoxic T lymphocyte-associated antigen 4 (CTLA4), interleukin (IL)-2/IL-21, IL-2 receptor A (IL-2RA; CD25) and Eos (also known as Ikaros family zinc finger 4; IKZF4), as well as the human leukocyte antigen (HLA) region. We also find association evidence for regions containing genes expressed in the hair follicle itself (PRDX5 and STX17). A region of strong association resides within the ULBP (cytomegalovirus UL16-binding protein) gene cluster on chromosome 6q25.1, encoding activating ligands of the natural killer cell receptor NKG2D that have not previously been implicated in an autoimmune disease. By probing the role of ULBP3 in disease pathogenesis, we also show that its expression in lesional scalp from patients with AA is markedly upregulated in the hair follicle dermal sheath during active disease. This study provides evidence for the involvement of both innate and acquired immunity in the pathogenesis of AA. We have defined the genetic underpinnings of AA, placing it within the context of shared pathways among autoimmune diseases, and implicating a novel disease mechanism, the upregulation of ULBP ligands, in triggering autoimmunity.
Figures

References
-
- Safavi KH, Muller SA, Suman VJ, Moshell AN, Melton LJ., III Incidence of alopecia areata in Olmsted County, Minnesota, 1975 through 1989. Mayo Clin. Proc. 1995;70:628–633. - PubMed
-
- Gilhar A, et al. Transfer of alopecia areata in the human scalp graft/Prkdc(scid) (SCID) mouse system is characterized by a TH1 response. Clin. Immunol. 2003;106:181–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

