Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization
- PMID: 20596258
- PMCID: PMC2893129
- DOI: 10.1371/journal.pone.0011335
Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization
Abstract
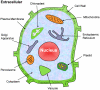
One of the fundamental goals in proteomics and cell biology is to identify the functions of proteins in various cellular organelles and pathways. Information of subcellular locations of proteins can provide useful insights for revealing their functions and understanding how they interact with each other in cellular network systems. Most of the existing methods in predicting plant protein subcellular localization can only cover three or four location sites, and none of them can be used to deal with multiplex plant proteins that can simultaneously exist at two, or move between, two or more different location sits. Actually, such multiplex proteins might have special biological functions worthy of particular notice. The present study was devoted to improve the existing plant protein subcellular location predictors from the aforementioned two aspects. A new predictor called "Plant-mPLoc" is developed by integrating the gene ontology information, functional domain information, and sequential evolutionary information through three different modes of pseudo amino acid composition. It can be used to identify plant proteins among the following 12 location sites: (1) cell membrane, (2) cell wall, (3) chloroplast, (4) cytoplasm, (5) endoplasmic reticulum, (6) extracellular, (7) Golgi apparatus, (8) mitochondrion, (9) nucleus, (10) peroxisome, (11) plastid, and (12) vacuole. Compared with the existing methods for predicting plant protein subcellular localization, the new predictor is much more powerful and flexible. Particularly, it also has the capacity to deal with multiple-location proteins, which is beyond the reach of any existing predictors specialized for identifying plant protein subcellular localization. As a user-friendly web-server, Plant-mPLoc is freely accessible at http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/. Moreover, for the convenience of the vast majority of experimental scientists, a step-by-step guide is provided on how to use the web-server to get the desired results. It is anticipated that the Plant-mPLoc predictor as presented in this paper will become a very useful tool in plant science as well as all the relevant areas.
Conflict of interest statement
Figures

References
-
- Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007;12:7–16. - PubMed
-
- Nakashima H, Nishikawa K. Discrimination of intracellular and extracellular proteins using amino acid composition and residue-pair frequencies. J Mol Biol. 1994;238:54–61. - PubMed
-
- Cedano J, Aloy P, P'erez-Pons JA, Querol E. Relation between amino acid composition and cellular location of proteins. J Mol Biol. 1997;266:594–600. - PubMed
-
- Chou KC, Elrod DW. Protein subcellular location prediction. Protein Engineering. 1999;12:107–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

