Isolation of Chlamydia pneumoniae from serum samples of the patients with acute coronary syndrome
- PMID: 20596362
- PMCID: PMC2894221
- DOI: 10.7150/ijms.7.181
Isolation of Chlamydia pneumoniae from serum samples of the patients with acute coronary syndrome
Abstract
Background: Limited body of evidence suggests that lipopolysaccharide of C. pneumoniae as well as C. pneumoniae-specific immune complexes can be detected and isolated from human serum. The aim of this study was to investigate the presence of viable elementary bodies of C.pneumoniae in serum samples of patients with acute coronary syndrome and healthy volunteers.
Material and methods: Serum specimens from 26 healthy volunteers and 56 patients with acute coronary syndrome were examined subsequently by serological (C.pneumoniae-specific IgA and IgG), PCR-based and bacteriological methods. Conventional, nested and TaqMan PCR were used to detect C.pneumoniae genetic markers (ompA and 16S rRNA) in DNA from serum specimens extracted with different methods. An alternative protocol which included culturing high-speed serum sediments in HL cells and further C.pneumoniae growth evaluation with immunofluorescence analysis and TaqMan PCR was established. Pellet fraction of PCR-positive serum specimens was also examined by immunoelectron microscopy.
Results: Best efficiency of final PCR product recovery from serum specimens has been shown with specific C. pneumoniae primers using phenol-chloroform DNA extraction protocol. TaqMan PCR analysis revealed that human serum of patients with acute coronary syndrome may contain genetic markers of C. pneumoniae with bacterial load range from 200 to 2000 copies/ml serum. However, reliability and reproducibility of TaqMan PCR were poor for serum specimens with low bacterial copy number (<200 /ml). Combination of bacteriological, immunofluorescence and PCR- based protocols applied for the evaluating HL cells infected with serum sediments revealed that 21.0 % of the patients with acute coronary syndrome have viable forms C.pneumoniae in serum. The detection rate of C.pneumoniae in healthy volunteers was much lower (7.7%). Immunological profile of the patients did not match accurately C.pneumoniae detection rate in serum specimens. Elementary bodies of C.pneumoniae with typical ultrastructural characteristics were also identified in serum sediments using immunoelectron microscopy.
Conclusions: Viable forms C. pneumoniae with typical electron microscopic structure can be identified and isolated from serum specimens of the patients with acute coronary syndrome and some healthy volunteers. Increased detection rate of C. pneumoniae in serum among the patients with an acute coronary syndrome may contribute towards enhanced pro-inflammatory status in cardiovascular patients and development of secondary complications of atherosclerosis.
Keywords: Chlamydia pneumoniae; PCR; acute coronary syndrome; cultured cells; human serum.
Conflict of interest statement
Conflict of Interest: The authors have declared that no conflict of interest exists.
Figures




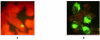
Similar articles
-
Comparison of a new quantitative ompA-based real-Time PCR TaqMan assay for detection of Chlamydia pneumoniae DNA in respiratory specimens with four conventional PCR assays.J Clin Microbiol. 2003 Feb;41(2):592-600. doi: 10.1128/JCM.41.2.592-600.2003. J Clin Microbiol. 2003. PMID: 12574252 Free PMC article.
-
Analytical sensitivity, reproducibility of results, and clinical performance of five PCR assays for detecting Chlamydia pneumoniae DNA in peripheral blood mononuclear cells.J Clin Microbiol. 2000 Jul;38(7):2622-7. doi: 10.1128/JCM.38.7.2622-2627.2000. J Clin Microbiol. 2000. PMID: 10878053 Free PMC article.
-
Comparison of five PCR assays for detecting Chlamydophila pneumoniae DNA.Microbiol Immunol. 2004;48(6):441-8. doi: 10.1111/j.1348-0421.2004.tb03534.x. Microbiol Immunol. 2004. PMID: 15215617
-
Reliability of nested PCR for detection of Chlamydia pneumoniae DNA in atheromas: results from a multicenter study applying standardized protocols.J Clin Microbiol. 2002 Dec;40(12):4428-34. doi: 10.1128/JCM.40.12.4428-4434.2002. J Clin Microbiol. 2002. PMID: 12454131 Free PMC article.
-
Failure to detect Chlamydia pneumoniae DNA in cerebral aneurysmal sac tissue with two different polymerase chain reaction methods.J Neurol Neurosurg Psychiatry. 2003 Jun;74(6):756-9. doi: 10.1136/jnnp.74.6.756. J Neurol Neurosurg Psychiatry. 2003. PMID: 12754346 Free PMC article. Review.
Cited by
-
Association of Chlamydia pneumoniae Infection With Atherosclerotic Plaque Formation.Glob J Health Sci. 2015 Sep 28;8(4):260-7. doi: 10.5539/gjhs.v8n4p260. Glob J Health Sci. 2015. PMID: 26573036 Free PMC article.
-
Lycopene Inhibits Propagation of Chlamydia Infection.Scientifica (Cairo). 2017;2017:1478625. doi: 10.1155/2017/1478625. Epub 2017 Aug 29. Scientifica (Cairo). 2017. PMID: 28948060 Free PMC article.
-
Detection of C. trachomatis in the serum of the patients with urogenital chlamydiosis.Biomed Res Int. 2013;2013:489489. doi: 10.1155/2013/489489. Epub 2013 Feb 13. Biomed Res Int. 2013. PMID: 23509729 Free PMC article.
-
Selective Pressure Promotes Tetracycline Resistance of Chlamydia Suis in Fattening Pigs.PLoS One. 2016 Nov 28;11(11):e0166917. doi: 10.1371/journal.pone.0166917. eCollection 2016. PLoS One. 2016. PMID: 27893834 Free PMC article.
-
Is there any relationship between Chlamydophila pneumoniae and coronary atherosclerosis among Iranians?Niger Med J. 2013 Jan;54(1):40-4. doi: 10.4103/0300-1652.108894. Niger Med J. 2013. PMID: 23661898 Free PMC article.
References
-
- Shor A. Chlamydia atherosclerosis lesion, discovery, diagnosis and treatment. Springer-Verlag. 2007.
-
- Stratton CW, Wheldon DB. Multiple sclerosis: an infectious syndrome involving Chlamydophila pneumoniae. Trends Microbiol. 2006 Nov;14(11):474–9. - PubMed
-
- Stallings TL. Association of Alzheimer's disease and Chlamydophila pneumoniae. J Infect. 2008 Jun;56(6):423–31. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

