Digital PCR on a SlipChip
- PMID: 20596567
- PMCID: PMC2948063
- DOI: 10.1039/c004521g
Digital PCR on a SlipChip
Abstract
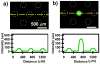
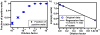
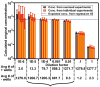
This paper describes a SlipChip to perform digital PCR in a very simple and inexpensive format. The fluidic path for introducing the sample combined with the PCR mixture was formed using elongated wells in the two plates of the SlipChip designed to overlap during sample loading. This fluidic path was broken up by simple slipping of the two plates that removed the overlap among wells and brought each well in contact with a reservoir preloaded with oil to generate 1280 reaction compartments (2.6 nL each) simultaneously. After thermal cycling, end-point fluorescence intensity was used to detect the presence of nucleic acid. Digital PCR on the SlipChip was tested quantitatively by using Staphylococcus aureus genomic DNA. As the concentration of the template DNA in the reaction mixture was diluted, the fraction of positive wells decreased as expected from the statistical analysis. No cross-contamination was observed during the experiments. At the extremes of the dynamic range of digital PCR the standard confidence interval determined using a normal approximation of the binomial distribution is not satisfactory. Therefore, statistical analysis based on the score method was used to establish these confidence intervals. The SlipChip provides a simple strategy to count nucleic acids by using PCR. It may find applications in research applications such as single cell analysis, prenatal diagnostics, and point-of-care diagnostics. SlipChip would become valuable for diagnostics, including applications in resource-limited areas after integration with isothermal nucleic acid amplification technologies and visual readout.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

