Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering
- PMID: 20597502
- PMCID: PMC2918669
- DOI: 10.1021/ja103867p
Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering
Abstract
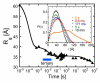
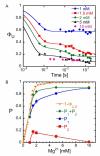
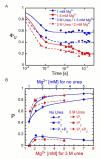
Ribozymes must fold into compact, native structures to function properly in the cell. The first step in forming the RNA tertiary structure is the neutralization of the phosphate charge by cations, followed by collapse of the unfolded molecules into more compact structures. The specificity of the collapse transition determines the structures of the folding intermediates and the folding time to the native state. However, the forces that enable specific collapse in RNA are not understood. Using time-resolved SAXS, we report that upon addition of 5 mM Mg(2+) to the Azoarcus group I ribozyme up to 80% of chains form compact structures in less than 1 ms. In 1 mM Mg(2+), the collapse transition produces extended structures that slowly approach the folded state, while > or = 1.5 mM Mg(2+) leads to an ensemble of random coils that fold with multistage kinetics. Increased flexibility of molecules in the intermediate ensemble correlates with a Mg(2+)-dependent increase in the fast folding population and a previously unobserved crossover in the collapse kinetics. Partial denaturation of the unfolded RNA with urea also increases the fraction of chains following the fast-folding pathway. These results demonstrate that the preferred collapse mechanism depends on the extent of Mg(2+)-dependent charge neutralization and that non-native interactions within the unfolded ensemble contribute to the heterogeneity of the ribozyme folding pathways at the very earliest stages of tertiary structure formation.
Figures



Similar articles
-
Metal ion dependence of cooperative collapse transitions in RNA.J Mol Biol. 2009 Oct 30;393(3):753-64. doi: 10.1016/j.jmb.2009.08.044. Epub 2009 Aug 25. J Mol Biol. 2009. PMID: 19712681 Free PMC article.
-
Effects of Preferential Counterion Interactions on the Specificity of RNA Folding.J Phys Chem Lett. 2018 Oct 4;9(19):5726-5732. doi: 10.1021/acs.jpclett.8b02086. Epub 2018 Sep 18. J Phys Chem Lett. 2018. PMID: 30211556 Free PMC article.
-
RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme.J Mol Biol. 2005 Nov 11;353(5):1199-209. doi: 10.1016/j.jmb.2005.09.015. Epub 2005 Sep 23. J Mol Biol. 2005. PMID: 16214167
-
Compact intermediates in RNA folding.Annu Rev Biophys. 2010;39:61-77. doi: 10.1146/annurev.biophys.093008.131334. Annu Rev Biophys. 2010. PMID: 20192764 Free PMC article. Review.
-
Folding and activity of the hammerhead ribozyme.Chembiochem. 2002 Aug 2;3(8):690-700. doi: 10.1002/1439-7633(20020802)3:8<690::AID-CBIC690>3.0.CO;2-C. Chembiochem. 2002. PMID: 12203967 Review.
Cited by
-
Toward a molecular understanding of RNA remodeling by DEAD-box proteins.RNA Biol. 2013 Jan;10(1):44-55. doi: 10.4161/rna.22210. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995827 Free PMC article. Review.
-
Sub-millisecond time-resolved SAXS using a continuous-flow mixer and X-ray microbeam.J Synchrotron Radiat. 2013 Nov;20(Pt 6):820-5. doi: 10.1107/S0909049513021833. Epub 2013 Oct 1. J Synchrotron Radiat. 2013. PMID: 24121320 Free PMC article.
-
Molecular crowding overcomes the destabilizing effects of mutations in a bacterial ribozyme.Nucleic Acids Res. 2015 Jan;43(2):1170-6. doi: 10.1093/nar/gku1335. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25541198 Free PMC article.
-
Methods for SAXS-based structure determination of biomolecular complexes.Adv Mater. 2014 Dec 10;26(46):7902-10. doi: 10.1002/adma.201304475. Epub 2014 May 30. Adv Mater. 2014. PMID: 24888261 Free PMC article.
-
Role of ion valence in the submillisecond collapse and folding of a small RNA domain.Biochemistry. 2013 Mar 5;52(9):1539-46. doi: 10.1021/bi3016636. Epub 2013 Feb 21. Biochemistry. 2013. PMID: 23398396 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

