Screening the human exome: a comparison of whole genome and whole transcriptome sequencing
- PMID: 20598109
- PMCID: PMC2898068
- DOI: 10.1186/gb-2010-11-5-r57
Screening the human exome: a comparison of whole genome and whole transcriptome sequencing
Abstract
Background: There is considerable interest in the development of methods to efficiently identify all coding variants present in large sample sets of humans. There are three approaches possible: whole-genome sequencing, whole-exome sequencing using exon capture methods, and RNA-Seq. While whole-genome sequencing is the most complete, it remains sufficiently expensive that cost effective alternatives are important.
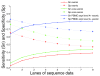
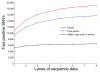
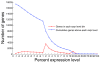
Results: Here we provide a systematic exploration of how well RNA-Seq can identify human coding variants by comparing variants identified through high coverage whole-genome sequencing to those identified by high coverage RNA-Seq in the same individual. This comparison allowed us to directly evaluate the sensitivity and specificity of RNA-Seq in identifying coding variants, and to evaluate how key parameters such as the degree of coverage and the expression levels of genes interact to influence performance. We find that although only 40% of exonic variants identified by whole genome sequencing were captured using RNA-Seq; this number rose to 81% when concentrating on genes known to be well-expressed in the source tissue. We also find that a high false positive rate can be problematic when working with RNA-Seq data, especially at higher levels of coverage.
Conclusions: We conclude that as long as a tissue relevant to the trait under study is available and suitable quality control screens are implemented, RNA-Seq is a fast and inexpensive alternative approach for finding coding variants in genes with sufficiently high expression levels.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

