Orthology confers intron position conservation
- PMID: 20598118
- PMCID: PMC2996940
- DOI: 10.1186/1471-2164-11-412
Orthology confers intron position conservation
Abstract
Background: With the wealth of genomic data available it has become increasingly important to assign putative protein function through functional transfer between orthologs. Therefore, correct elucidation of the evolutionary relationships among genes is a critical task, and attempts should be made to further improve the phylogenetic inference by adding relevant discriminating features. It has been shown that introns can maintain their position over long evolutionary timescales. For this reason, it could be possible to use conservation of intron positions as a discriminating factor when assigning orthology. Therefore, we wanted to investigate whether orthologs have a higher degree of intron position conservation (IPC) compared to non-orthologous sequences that are equally similar in sequence.
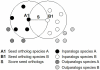
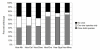
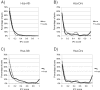
Results: To this end, we developed a new score for IPC and applied it to ortholog groups between human and six other species. For comparison, we also gathered the closest non-orthologs, meaning sequences close in sequence space, yet falling just outside the ortholog cluster. We found that ortholog-ortholog gene pairs on average have a significantly higher degree of IPC compared to ortholog-closest non-ortholog pairs. Also pairs of inparalogs were found to have a higher IPC score than inparalog-closest non-inparalog pairs. We verified that these differences can not simply be attributed to the generally higher sequence identity of the ortholog-ortholog and the inparalog-inparalog pairs. Furthermore, we analyzed the agreement between IPC score and the ortholog score assigned by the InParanoid algorithm, and found that it was consistently high for all species comparisons. In a minority of cases, the IPC and InParanoid score ranked inparalogs differently. These represent cases where sequence and intron position divergence are discordant. We further analyzed the discordant clusters to identify any possible preference for protein functions by looking for enriched GO terms and Pfam protein domains. They were enriched for functions important for multicellularity, which implies a connection between shifts in intronic structure and the origin of multicellularity.
Conclusions: We conclude that orthologous genes tend to have more conserved intron positions compared to non-orthologous genes. As a consequence, our IPC score is useful as an additional discriminating factor when assigning orthology.
Figures



Similar articles
-
InParanoid 6: eukaryotic ortholog clusters with inparalogs.Nucleic Acids Res. 2008 Jan;36(Database issue):D263-6. doi: 10.1093/nar/gkm1020. Epub 2007 Nov 30. Nucleic Acids Res. 2008. PMID: 18055500 Free PMC article.
-
Automatic clustering of orthologs and inparalogs shared by multiple proteomes.Bioinformatics. 2006 Jul 15;22(14):e9-15. doi: 10.1093/bioinformatics/btl213. Bioinformatics. 2006. PMID: 16873526
-
Intron position conservation across eukaryotic lineages in tubulin genes.Front Biosci. 2005 Sep 1;10:2412-9. doi: 10.2741/1706. Front Biosci. 2005. PMID: 15970504
-
Analysis of evolution of exon-intron structure of eukaryotic genes.Brief Bioinform. 2005 Jun;6(2):118-34. doi: 10.1093/bib/6.2.118. Brief Bioinform. 2005. PMID: 15975222 Review.
-
Spliceosomal introns as tools for genomic and evolutionary analysis.Nucleic Acids Res. 2008 Mar;36(5):1703-12. doi: 10.1093/nar/gkn012. Epub 2008 Feb 7. Nucleic Acids Res. 2008. PMID: 18263615 Free PMC article. Review.
Cited by
-
Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs.PLoS Comput Biol. 2012;8(5):e1002514. doi: 10.1371/journal.pcbi.1002514. Epub 2012 May 17. PLoS Comput Biol. 2012. PMID: 22615551 Free PMC article.
-
The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth.J Exp Bot. 2012 Sep;63(15):5417-35. doi: 10.1093/jxb/ers194. Epub 2012 Aug 9. J Exp Bot. 2012. PMID: 22888130 Free PMC article.
-
Positive Selection Linked with Generation of Novel Mammalian Dentition Patterns.Genome Biol Evol. 2016 Sep 11;8(9):2748-59. doi: 10.1093/gbe/evw200. Genome Biol Evol. 2016. PMID: 27613398 Free PMC article.
-
Network analysis of functional genomics data: application to avian sex-biased gene expression.ScientificWorldJournal. 2012;2012:130491. doi: 10.1100/2012/130491. Epub 2012 Dec 23. ScientificWorldJournal. 2012. PMID: 23319882 Free PMC article.
-
Taxonomic Distribution and Molecular Evolution of Mytilectins.Mar Drugs. 2023 Nov 27;21(12):614. doi: 10.3390/md21120614. Mar Drugs. 2023. PMID: 38132935 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

