Targeting breast cancer stem cells
- PMID: 20599450
- PMCID: PMC3023958
- DOI: 10.1016/j.molonc.2010.06.005
Targeting breast cancer stem cells
Abstract
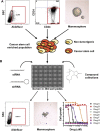
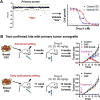
The cancer stem cell (CSC) hypothesis postulates that tumors are maintained by a self-renewing CSC population that is also capable of differentiating into non-self-renewing cell populations that constitute the bulk of the tumor. Although, the CSC hypothesis does not directly address the cell of origin of cancer, it is postulated that tissue-resident stem or progenitor cells are the most common targets of transformation. Clinically, CSCs are predicted to mediate tumor recurrence after chemo- and radiation-therapy due to the relative inability of these modalities to effectively target CSCs. If this is the case, then CSC must be efficiently targeted to achieve a true cure. Similarities between normal and malignant stem cells, at the levels of cell-surface proteins, molecular pathways, cell cycle quiescence, and microRNA signaling present challenges in developing CSC-specific therapeutics. Approaches to targeting CSCs include the development of agents targeting known stem cell regulatory pathways as well as unbiased high-throughput siRNA or small molecule screening. Based on studies of pathways present in normal stem cells, recent work has identified potential "Achilles heals" of CSC, whereas unbiased screening provides opportunities to identify new pathways utilized by CSC as well as develop potential therapeutic agents. Here, we review both approaches and their potential to effectively target breast CSC.
Copyright © 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
References
-
- Abratt, R.P. , Brune, D. , Dimopoulos, M.A. , Kliment, J. , Breza, J. , Selvaggi, F.P. , Beuzeboc, P. , Demkow, T. , Oudard, S. , 2004. Randomised phase III study of intravenous vinorelbine plus hormone therapy versus hormone therapy alone in hormone-refractory prostate cancer. Ann. Oncol. 15, 1613–1621. - PubMed
-
- Agis, H. , Jaeger, E. , Doninger, B. , Sillaber, C. , Marosi, C. , Drach, J. , Schwarzinger, I. , Valent, P. , Oehler, L. , 2006. In vivo effects of imatinib mesylate on human haematopoietic progenitor cells. Eur. J. Clin. Invest. 36, 402–408. - PubMed
-
- Bao, S. , Wu, Q. , McLendon, R.E. , Hao, Y. , Shi, Q. , Hjelmeland, A.B. , Dewhirst, M.W. , Bigner, D.D. , Rich, J.N. , 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444, 756–760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

