Action of the chaperonin GroEL/ES on a non-native substrate observed with single-molecule FRET
- PMID: 20600107
- PMCID: PMC2927214
- DOI: 10.1016/j.jmb.2010.06.050
Action of the chaperonin GroEL/ES on a non-native substrate observed with single-molecule FRET
Abstract
The double ring-shaped chaperonin GroEL binds a wide range of non-native polypeptides within its central cavity and, together with its cofactor GroES, assists their folding in an ATP-dependent manner. The conformational cycle of GroEL/ES has been studied extensively but little is known about how the environment in the central cavity affects substrate conformation. Here, we use the von Hippel-Lindau tumor suppressor protein VHL as a model substrate for studying the action of the GroEL/ES system on a bound polypeptide. Fluorescent labeling of pairs of sites on VHL for fluorescence (Förster) resonant energy transfer (FRET) allows VHL to be used to explore how GroEL binding and GroEL/ES/nucleotide binding affect the substrate conformation. On average, upon binding to GroEL, all pairs of labeling sites experience compaction relative to the unfolded protein while single-molecule FRET distributions show significant heterogeneity. Upon addition of GroES and ATP to close the GroEL cavity, on average further FRET increases occur between the two hydrophobic regions of VHL, accompanied by FRET decreases between the N- and C-termini. This suggests that ATP- and GroES-induced confinement within the GroEL cavity remodels bound polypeptides by causing expansion (or racking) of some regions and compaction of others, most notably, the hydrophobic core. However, single-molecule observations of the specific FRET changes for individual proteins at the moment of ATP/GroES addition reveal that a large fraction of the population shows the opposite behavior; that is, FRET decreases between the hydrophobic regions and FRET increases for the N- and C-termini. Our time-resolved single-molecule analysis reveals the underlying heterogeneity of the action of GroES/EL on a bound polypeptide substrate, which might arise from the random nature of the specific binding to the various identical subunits of GroEL, and might help explain why multiple rounds of binding and hydrolysis are required for some chaperonin substrates.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
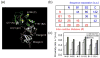
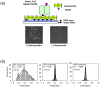
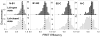


Similar articles
-
Application of fluorescence resonance energy transfer to the GroEL-GroES chaperonin reaction.Methods. 2001 Jul;24(3):278-88. doi: 10.1006/meth.2001.1188. Methods. 2001. PMID: 11403576 Free PMC article. Review.
-
Active cage mechanism of chaperonin-assisted protein folding demonstrated at single-molecule level.J Mol Biol. 2014 Jul 29;426(15):2739-54. doi: 10.1016/j.jmb.2014.04.018. Epub 2014 May 6. J Mol Biol. 2014. PMID: 24816391
-
Substrate polypeptide presents a load on the apical domains of the chaperonin GroEL.Proc Natl Acad Sci U S A. 2004 Oct 19;101(42):15005-12. doi: 10.1073/pnas.0406132101. Epub 2004 Oct 12. Proc Natl Acad Sci U S A. 2004. PMID: 15479763 Free PMC article.
-
Chaperonin-Assisted Protein Folding: Relative Population of Asymmetric and Symmetric GroEL:GroES Complexes.J Mol Biol. 2015 Jun 19;427(12):2244-55. doi: 10.1016/j.jmb.2015.04.009. Epub 2015 Apr 23. J Mol Biol. 2015. PMID: 25912285
-
Protein folding assisted by the GroEL/GroES chaperonin system.Biochemistry (Mosc). 1998 Apr;63(4):374-81. Biochemistry (Mosc). 1998. PMID: 9556520 Review.
Cited by
-
pVHL-mediated SMAD3 degradation suppresses TGF-β signaling.J Cell Biol. 2022 Jan 3;221(1):e202012097. doi: 10.1083/jcb.202012097. Epub 2021 Dec 3. J Cell Biol. 2022. PMID: 34860252 Free PMC article.
-
smFRET Detection of Cis and Trans DNA Interactions by the BfiI Restriction Endonuclease.J Phys Chem B. 2023 Jul 27;127(29):6470-6478. doi: 10.1021/acs.jpcb.3c03269. Epub 2023 Jul 15. J Phys Chem B. 2023. PMID: 37452775 Free PMC article.
-
Nucleotide-induced conformational changes of tetradecameric GroEL mapped by H/D exchange monitored by FT-ICR mass spectrometry.Sci Rep. 2013;3:1247. doi: 10.1038/srep01247. Epub 2013 Feb 13. Sci Rep. 2013. PMID: 23409238 Free PMC article.
-
Live-cell imaging of single receptor composition using zero-mode waveguide nanostructures.Nano Lett. 2012 Jul 11;12(7):3690-4. doi: 10.1021/nl301480h. Epub 2012 Jun 8. Nano Lett. 2012. PMID: 22668081 Free PMC article.
-
Functional Subunits of Eukaryotic Chaperonin CCT/TRiC in Protein Folding.J Amino Acids. 2011;2011:843206. doi: 10.4061/2011/843206. Epub 2011 Jul 2. J Amino Acids. 2011. PMID: 22312474 Free PMC article.
References
-
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. - PubMed
-
- Dunn AY, Melville MW, Frydman J. Review: Cellular Substrates of the Eukaryotic Chaperonin TRiC/CCT. Journal of Structural Biology. 2001;135:176–184. - PubMed
-
- Fenton WA, Horwich AL. Chaperonin-mediated protein folding: fate of substrate polypeptide. Q Rev Biophys. 2003;36:229–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

