Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus
- PMID: 20600115
- PMCID: PMC3050670
- DOI: 10.1016/j.jmb.2010.06.042
Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus
Abstract
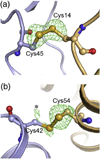
The human immunodeficiency virus type 1 capsid is modeled as a fullerene cone that is composed of approximately 250 hexamers and 12 pentamers of the viral CA protein. Structures of CA hexamers have been difficult to obtain because the hexamer-stabilizing interactions are inherently weak, and CA tends to spontaneously assemble into capsid-like particles. Here, we describe a two-step biochemical strategy to obtain soluble CA hexamers for crystallization. First, the hexamer was stabilized by engineering disulfide cross-links (either A14C/E45C or A42C/T54C) between the N-terminal domains of adjacent subunits. Second, the cross-linked hexamers were prevented from polymerizing further into hyperstable capsid-like structures by mutations (W184A and M185A) that interfered with dimeric association between the C-terminal domains that link adjacent hexamers. The structures of two different cross-linked CA hexamers were nearly identical, and we combined the non-mutated portions of the structures to generate an atomic resolution model for the native hexamer. This hybrid approach for structure determination should be applicable to other viral capsomers and protein-protein complexes in general.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures




References
-
- Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. - PubMed
-
- Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. - PubMed
-
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. - PubMed
-
- Mortuza GB, Haire LF, Stevens A, Smerdon SJ, Stoye JP, Taylor IA. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

