Conantokins inhibit NMDAR-dependent calcium influx in developing rat hippocampal neurons in primary culture with resulting effects on CREB phosphorylation
- PMID: 20600930
- PMCID: PMC2923249
- DOI: 10.1016/j.mcn.2010.06.007
Conantokins inhibit NMDAR-dependent calcium influx in developing rat hippocampal neurons in primary culture with resulting effects on CREB phosphorylation
Abstract
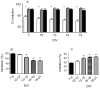
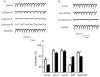
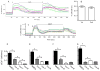
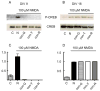
The effects of conantokin (con)-G, con-R[1-17], and con-T on ion flow through N-methyl-D-aspartate receptor (NMDAR) ion channels were determined in cultured primary rat hippocampal neurons. The potency of con-G diminished, whereas inhibition by con-R[1-17] and con-T did not change, as the neurons matured. Con-G, con-R[1-17], and con-T effectively diminished NMDA-induced Ca(2+) influx into the cells. A similar age-dependent decrease in con-G-mediated inhibition of the amplitude of spontaneous excitatory postsynaptic currents (sEPSCs) was observed, compared to con-R[1-17] and con-T. The effects of the conantokins on NMDA-induced cAMP response element-binding protein (CREB) phosphorylation in immature (DIV 9) and mature (DIV 16) neurons showed that, at DIV 9, con-G, con-R[1-17], and con-T inhibited NMDA-mediated P-CREB levels, whereas in DIV 16 neurons the conantokins did not inhibit overall levels of NMDA-induced P-CREB. In contrast, P-CREB levels were enhanced through inhibition of the protein phosphatases, PP1 and PP2B (calcineurin). This ability of conantokins to sustain CREB phosphorylation can thus enhance neuronal survival and plasticity.
Copyright 2010 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Alex AB, Baucum AJ, Wilcox KS. Effect of Conantokin G on NMDA receptor-mediated spontaneous EPSCs in cultured cortical neurons. J Neurophysiol. 2006;96:1084–1092. - PubMed
-
- Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M. 5-phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg Med Chem Lett. 2002;12:1099–1102. - PubMed
-
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. - PubMed
-
- Barton ME, White HS, Wilcox KS. The effect of CGX-1007 and CI-1041, novel NMDA receptor antagonists, on NMDA receptor-mediated EPSCs. Epilepsy Res. 2004;59:13–24. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

