Influence of the second amino acid on recombinant protein expression
- PMID: 20600944
- PMCID: PMC2952673
- DOI: 10.1016/j.pep.2010.06.005
Influence of the second amino acid on recombinant protein expression
Abstract
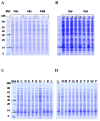
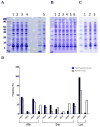
Factors affecting protein expression have been intensely studied to the benefit of recombinant protein production. Through mutational analysis at the +2 amino acid position of recombinant Igα, we examined the effect of all 20 amino acids on protein expression. The results showed that amino acids at the +2 position affected 10-fold in the recombinant protein expression. Specifically, Ala, Cys, Pro, Ser, Thr, and Lys at the +2 position resulted in significantly higher expression of recombinant Igα than other amino acids, while Met, His and Glu resulted in greatly reduced protein expression. This expression difference depended on the amino acid instead of their codon usage. Consistent with the mutational results, a statistically significant enrichment in Ala and Ser at the +2 position was observed among highly expressed Escherichia coli genes. This work suggests a general approach to enhance protein expression by incorporating an Ala or Ser after the initiation codon.
Published by Elsevier Inc.
Figures







Similar articles
-
Structure and binding studies of proliferating cell nuclear antigen from Leishmania donovani.Biochim Biophys Acta Proteins Proteom. 2017 Nov;1865(11 Pt A):1395-1405. doi: 10.1016/j.bbapap.2017.08.011. Epub 2017 Aug 24. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 28844736
-
The amino acid sequence of human chorionic gonadotropin. The alpha subunit and beta subunit.J Biol Chem. 1975 Jul 10;250(13):5247-58. J Biol Chem. 1975. PMID: 1150658
-
Complete amino acid sequence of copper-zinc superoxide dismutase from Drosophila melanogaster.Arch Biochem Biophys. 1985 Sep;241(2):577-89. doi: 10.1016/0003-9861(85)90583-1. Arch Biochem Biophys. 1985. PMID: 3929689
-
The primary structure of human liver manganese superoxide dismutase.J Biol Chem. 1984 Oct 25;259(20):12595-601. J Biol Chem. 1984. PMID: 6386798
-
Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering.Biotechnol Annu Rev. 2007;13:27-42. doi: 10.1016/S1387-2656(07)13002-7. Biotechnol Annu Rev. 2007. PMID: 17875472 Review.
Cited by
-
The genotype-phenotype landscape of an allosteric protein.Mol Syst Biol. 2021 Mar;17(3):e10179. doi: 10.15252/msb.202010179. Mol Syst Biol. 2021. PMID: 33784029 Free PMC article.
-
Optimized Heterologous Expression and Efficient Purification of a New TRAIL-Based Antitumor Fusion Protein SRH-DR5-B with Dual VEGFR2 and DR5 Receptor Specificity.Int J Mol Sci. 2022 May 24;23(11):5860. doi: 10.3390/ijms23115860. Int J Mol Sci. 2022. PMID: 35682540 Free PMC article.
-
Evaluation of GFP reporter utility for analysis of transcriptional slippage during gene expression.Microb Cell Fact. 2018 Sep 21;17(1):150. doi: 10.1186/s12934-018-0999-3. Microb Cell Fact. 2018. PMID: 30241530 Free PMC article.
-
N-Terminal Amino Acid Affects the Translation Efficiency at Lower Temperatures in a Reconstituted Protein Synthesis System.Int J Mol Sci. 2024 May 12;25(10):5264. doi: 10.3390/ijms25105264. Int J Mol Sci. 2024. PMID: 38791303 Free PMC article.
-
Adenine Enrichment at the Fourth CDS Residue in Bacterial Genes Is Consistent with Error Proofing for +1 Frameshifts.Mol Biol Evol. 2017 Dec 1;34(12):3064-3080. doi: 10.1093/molbev/msx223. Mol Biol Evol. 2017. PMID: 28961919 Free PMC article.
References
-
- Shine J, Dalgarno L. Terminal-sequence analysis of bacterial ribosomal RNA. Correlation between the 3′-terminal-polypyrimidine sequence of 16-S RNA and translational specificity of the ribosome. Eur J Biochem. 1975;57:221–30. - PubMed
-
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. - PubMed
-
- Gren EJ. Recognition of messenger RNA during translational initiation in Escherichia coli. Biochimie. 1984;66:1–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

