iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution
- PMID: 20601959
- PMCID: PMC3000544
- DOI: 10.1038/nsmb.1838
iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution
Abstract
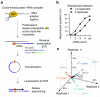
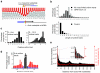
In the nucleus of eukaryotic cells, nascent transcripts are associated with heterogeneous nuclear ribonucleoprotein (hnRNP) particles that are nucleated by hnRNP C. Despite their abundance, however, it remained unclear whether these particles control pre-mRNA processing. Here, we developed individual-nucleotide resolution UV cross-linking and immunoprecipitation (iCLIP) to study the role of hnRNP C in splicing regulation. iCLIP data show that hnRNP C recognizes uridine tracts with a defined long-range spacing consistent with hnRNP particle organization. hnRNP particles assemble on both introns and exons but remain generally excluded from splice sites. Integration of transcriptome-wide iCLIP data and alternative splicing profiles into an 'RNA map' indicates how the positioning of hnRNP particles determines their effect on the inclusion of alternative exons. The ability of high-resolution iCLIP data to provide insights into the mechanism of this regulation holds promise for studies of other higher-order ribonucleoprotein complexes.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 089701/WT_/Wellcome Trust/United Kingdom
- MC_U105185858/MRC_/Medical Research Council/United Kingdom
- BB/E01075X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U.1051.04.028.00001.01 (85858)/MRC_/Medical Research Council/United Kingdom
- 206726/ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

