IRF4 variants have age-specific effects on nevus count and predispose to melanoma
- PMID: 20602913
- PMCID: PMC2896771
- DOI: 10.1016/j.ajhg.2010.05.017
IRF4 variants have age-specific effects on nevus count and predispose to melanoma
Abstract
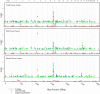
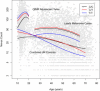
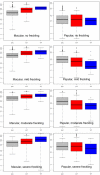
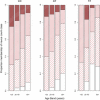
High melanocytic nevus count is a strong predictor of melanoma risk. A GWAS of nevus count in Australian adolescent twins identified an association of nevus count with the interferon regulatory factor 4 gene (IRF4 [p = 6 x 10(-9)]). There was a strong genotype-by-age interaction, which was replicated in independent UK samples of adolescents and adults. The rs12203592(*)T allele was associated with high nevus counts and high freckling scores in adolescents, but with low nevus counts and high freckling scores in adults. The rs12203592(*)T increased counts of flat (compound and junctional) nevi in Australian adolescent twins, but decreased counts of raised (intradermal) nevi. In combined analysis of melanoma case-control data from Australia, the UK, and Sweden, the rs12203592(*)C allele was associated with melanoma (odds ratio [OR] 1.15, p = 4 x 10(-3)), most significantly on the trunk (OR = 1.33, p = 2.5 x 10(-5)). The melanoma association was corroborated in a GWAS performed by the GenoMEL consortium for an adjacent SNP, rs872071 (rs872071(*)T: OR 1.14, p = 0.0035; excluding Australian, the UK, and Swedish samples typed at rs12203592: OR 1.08, p = 0.08).
Copyright 2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Czene K., Lichtenstein P., Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int. J. Cancer. 2002;99:260–266. - PubMed
-
- Begg C.B., Hummer A., Mujumdar U., Armstrong B.K., Kricker A., Marrett L.D., Millikan R.C., Gruber S.B., Anton-Culver H., Klotz J.B., GEM Study Group Familial aggregation of melanoma risks in a large population-based sample of melanoma cases. Cancer Causes Control. 2004;15:957–965. - PubMed
-
- Do K.A., Aitken J.F., Green A.C., Martin N.G. Analysis of melanoma onset: assessing familial aggregation by using estimating equations and fitting variance components via Bayesian random effects models. Twin Res. 2004;7:98–113. - PubMed
-
- Shekar S.N., Duffy D.L., Youl P., Baxter A.J., Kvaskoff M., Whiteman D.C., Green A.C., Hughes M.C., Hayward N.K., Coates M., Martin N.G. A population-based study of Australian twins with melanoma suggests a strong genetic contribution to liability. J. Invest. Dermatol. 2009;129:2211–2219. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

