Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes
- PMID: 20603003
- PMCID: PMC2935685
- DOI: 10.1016/j.cell.2010.05.007
Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes
Abstract
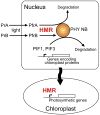
Light plays a profound role in plant development, yet how photoreceptor excitation directs phenotypic plasticity remains elusive. One of the earliest effects of light is the regulated translocation of the red/far-red photoreceptors, phytochromes, from the cytoplasm to subnuclear foci called phytochrome nuclear bodies. The function of these nuclear bodies is unknown. We report the identification of hemera, a seedling lethal mutant of Arabidopsis with altered phytochrome nuclear body patterns. hemera mutants are impaired in all phytochrome responses examined, including proteolysis of phytochrome A and phytochrome-interacting transcription factors. HEMERA was identified previously as pTAC12, a component of a plastid complex associated with transcription. Here, we show that HEMERA has a function in the nucleus, where it acts specifically in phytochrome signaling, is predicted to be structurally similar to the multiubiquitin-binding protein, RAD23, and can partially rescue yeast rad23mutants. Together, these results implicate phytochrome nuclear bodies as sites of proteolysis.
Figures
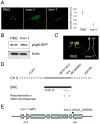
 ), hmr-2 (●), PBG (
), hmr-2 (●), PBG ( ), hmr-1 (
), hmr-1 ( ), and phyB-5 (
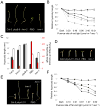
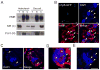
), and phyB-5 ( ) seedlings under different fluence of Rc and dark conditions. (C) EOD-FR responses of Col-0, hmr-2, phyB-9, PBG, and hmr-1. Filled columns represent hypocotyl lengths of 4-day old seedlings under 8-hour day/16-hour night; open columns represent hypocotyl lengths of 4-day old seedlings under the same short day conditions with an additional 15 min FR treatment at the end of the day. Red columns represent the percentage of increase in hypocotyl length of the treated seedlings compared to untreated seedlings. (D) Cotyledon opening responses for VLFR measurement. Cotyledon images of Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown under hourly 3 min 1 μmol m-2 sec-1 FR pulse for 4 days. (E) Images of 4-day old Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown in 1 μmol m-2 sec-1 FR light for 4 days. (F) Fluence response curves for FRc. Relative hypocotyl length of 4-day old Col-0 (◆), phyA-211 (■), hmr-2 (❄), PBG (
) seedlings under different fluence of Rc and dark conditions. (C) EOD-FR responses of Col-0, hmr-2, phyB-9, PBG, and hmr-1. Filled columns represent hypocotyl lengths of 4-day old seedlings under 8-hour day/16-hour night; open columns represent hypocotyl lengths of 4-day old seedlings under the same short day conditions with an additional 15 min FR treatment at the end of the day. Red columns represent the percentage of increase in hypocotyl length of the treated seedlings compared to untreated seedlings. (D) Cotyledon opening responses for VLFR measurement. Cotyledon images of Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown under hourly 3 min 1 μmol m-2 sec-1 FR pulse for 4 days. (E) Images of 4-day old Col-0, phyA-211, hmr-2, PBG, and hmr-1 seedlings grown in 1 μmol m-2 sec-1 FR light for 4 days. (F) Fluence response curves for FRc. Relative hypocotyl length of 4-day old Col-0 (◆), phyA-211 (■), hmr-2 (❄), PBG ( ), and hmr-1 (
), and hmr-1 ( ) seedlings grown under different fluence of FRc and dark conditions. Error bars represent standard error.
) seedlings grown under different fluence of FRc and dark conditions. Error bars represent standard error.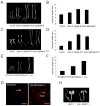

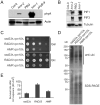
References
-
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
-
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

