Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2
- PMID: 20603015
- PMCID: PMC3710700
- DOI: 10.1016/j.cell.2010.06.021
Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2
Abstract
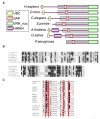
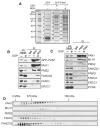
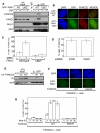
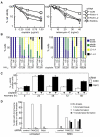
DNA interstrand crosslinks (ICLs) are highly toxic because they block the progression of replisomes. The Fanconi Anemia (FA) proteins, encoded by genes that are mutated in FA, are important for repair of ICLs. The FA core complex catalyzes the monoubiquitination of FANCD2, and this event is essential for several steps of ICL repair. However, how monoubiquitination of FANCD2 promotes ICL repair at the molecular level is unknown. Here, we describe a highly conserved protein, KIAA1018/MTMR15/FAN1, that interacts with, and is recruited to sites of DNA damage by, the monoubiquitinated form of FANCD2. FAN1 exhibits endonuclease activity toward 5' flaps and has 5' exonuclease activity, and these activities are mediated by an ancient VRR_nuc domain. Depletion of FAN1 from human cells causes hypersensitivity to ICLs, defects in ICL repair, and genome instability. These data at least partly explain how ubiquitination of FANCD2 promotes DNA repair.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures


Comment in
-
DNA repair has a new FAN1 club.Mol Cell. 2010 Jul 30;39(2):167-9. doi: 10.1016/j.molcel.2010.07.010. Mol Cell. 2010. PMID: 20670886
-
DNA repair: a new fan of the Fanconi anaemia pathway.Nat Rev Mol Cell Biol. 2010 Sep;11(9):603. doi: 10.1038/nrm2958. Epub 2010 Aug 11. Nat Rev Mol Cell Biol. 2010. PMID: 20700143 No abstract available.
References
-
- Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends Genet. 2008;24:70–76. - PubMed
-
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

