TIF1gamma controls erythroid cell fate by regulating transcription elongation
- PMID: 20603019
- PMCID: PMC3072682
- DOI: 10.1016/j.cell.2010.05.028
TIF1gamma controls erythroid cell fate by regulating transcription elongation
Abstract
Recent genome-wide studies have demonstrated that pausing of RNA polymerase II (Pol II) occurred on many vertebrate genes. By genetic studies in the zebrafish tif1gamma mutant moonshine we found that loss of function of Pol II-associated factors PAF or DSIF rescued erythroid gene transcription in tif1gamma-deficient animals. Biochemical analysis established physical interactions among TIF1gamma, the blood-specific SCL transcription complex, and the positive elongation factors p-TEFb and FACT. Chromatin immunoprecipitation assays in human CD34(+) cells supported a TIF1gamma-dependent recruitment of positive elongation factors to erythroid genes to promote transcription elongation by counteracting Pol II pausing. Our study establishes a mechanism for regulating tissue cell fate and differentiation through transcription elongation.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
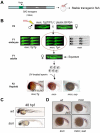
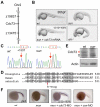

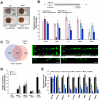


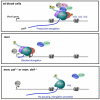
Comment in
-
Moonshine illuminates a developmental role for regulated transcription elongation.Dev Cell. 2010 Jul 20;19(1):9-10. doi: 10.1016/j.devcel.2010.07.003. Dev Cell. 2010. PMID: 20643345 Free PMC article.
References
-
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. - PubMed
-
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. - PubMed
-
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

