The cytoplasmic adaptor protein Dok7 activates the receptor tyrosine kinase MuSK via dimerization
- PMID: 20603078
- PMCID: PMC2917201
- DOI: 10.1016/j.molcel.2010.06.007
The cytoplasmic adaptor protein Dok7 activates the receptor tyrosine kinase MuSK via dimerization
Abstract
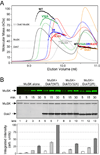
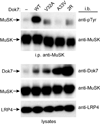
Formation of the vertebrate neuromuscular junction requires, among others proteins, Agrin, a neuronally derived ligand, and the following muscle proteins: LRP4, the receptor for Agrin; MuSK, a receptor tyrosine kinase (RTK); and Dok7 (or Dok-7), a cytoplasmic adaptor protein. Dok7 comprises a pleckstrin-homology (PH) domain, a phosphotyrosine-binding (PTB) domain, and C-terminal sites of tyrosine phosphorylation. Unique among adaptor proteins recruited to RTKs, Dok7 is not only a substrate of MuSK, but also an activator of MuSK's kinase activity. Here, we present the crystal structure of the Dok7 PH-PTB domains in complex with a phosphopeptide representing the Dok7-binding site on MuSK. The structure and biochemical data reveal a dimeric arrangement of Dok7 PH-PTB that facilitates trans-autophosphorylation of the kinase activation loop. The structure provides the molecular basis for MuSK activation by Dok7 and for rationalizing several Dok7 loss-of-function mutations found in patients with congenital myasthenic syndromes.
2010 Elsevier Inc. All rights reserved.
Figures




References
-
- Beeson D, Webster R, Cossins J, Lashley D, Spearman H, Maxwell S, Slater CR, Newsom-Davis J, Palace J, Vincent A. Congenital myasthenic syndromes and the formation of the neuromuscular junction. Ann. NY Acad. Sci. 2008;1132:99–103. - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. - PubMed
-
- Burden SJ. Building the vertebrate neuromuscular synapse. J. Neurobiol. 2002;53:501–511. - PubMed
-
- Cai D, Dhe-Paganon S, Melendez PA, Lee J, Shoelson SE. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J. Biol. Chem. 2003;278:25323–25330. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

