Alternative splicing and muscular dystrophy
- PMID: 20603608
- PMCID: PMC3568746
- DOI: 10.4161/rna.7.4.12258
Alternative splicing and muscular dystrophy
Abstract
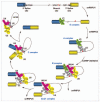
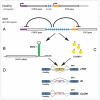
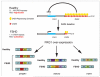
Alternative splicing of pre-mRNAs is a major contributor to proteomic diversity and to the control of gene expression in higher eukaryotic cells. For this reasons, alternative splicing is tightly regulated in different tissues and developmental stages and its disruption can lead to a wide range of human disorders. The aim of this review is to focus on the relevance of alternative splicing for muscle function and muscle disease. We begin by giving a brief overview of alternative splicing, muscle-specific gene expression and muscular dystrophy. Next, to illustrate these concepts we focus on two muscular dystrophy, myotonic muscular dystrophy and facioscapulohumeral muscular dystrophy, both associated to disruption of splicing regulation in muscle.
Figures


References
-
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. - PubMed
-
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. - PubMed
-
- Fu XD. Towards a splicing code. Cell. 2004;119:736–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
