Computational mutation scanning and drug resistance mechanisms of HIV-1 protease inhibitors
- PMID: 20604558
- PMCID: PMC2916083
- DOI: 10.1021/jp102546s
Computational mutation scanning and drug resistance mechanisms of HIV-1 protease inhibitors
Abstract
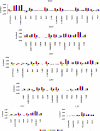
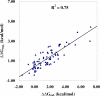
The drug resistance of various clinically available HIV-1 protease inhibitors has been studied using a new computational protocol, that is, computational mutation scanning (CMS), leading to valuable insights into the resistance mechanisms and structure-resistance correction of the HIV-1 protease inhibitors associated with a variety of active site and nonactive site mutations. By using the CMS method, the calculated mutation-caused shifts of the binding free energies linearly correlate very well with those derived from the corresponding experimental data, suggesting that the CMS protocol may be used as a generalized approach to predict drug resistance associated with amino acid mutations. Because it is essentially important for understanding the structure-resistance correlation and for structure-based drug design to develop an effective computational protocol for drug resistance prediction, the reasonable and computationally efficient CMS protocol for drug resistance prediction should be valuable for future structure-based design and discovery of antiresistance drugs in various therapeutic areas.
Figures
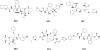




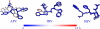
Similar articles
-
Prediction of HIV-1 protease inhibitor resistance using a protein-inhibitor flexible docking approach.Antivir Ther. 2005;10(1):157-66. Antivir Ther. 2005. PMID: 15751773
-
Genotype dependent QSAR for HIV-1 protease inhibition.J Med Chem. 2005 Mar 24;48(6):2115-20. doi: 10.1021/jm049596h. J Med Chem. 2005. PMID: 15771454
-
Computational Studies of a Mechanism for Binding and Drug Resistance in the Wild Type and Four Mutations of HIV-1 Protease with a GRL-0519 Inhibitor.Int J Mol Sci. 2016 May 27;17(6):819. doi: 10.3390/ijms17060819. Int J Mol Sci. 2016. PMID: 27240358 Free PMC article.
-
Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design.Curr Drug Targets Infect Disord. 2003 Dec;3(4):311-28. doi: 10.2174/1568005033481051. Curr Drug Targets Infect Disord. 2003. PMID: 14754432 Review.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
Cited by
-
Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.PLoS Pathog. 2018 Oct 22;14(10):e1007343. doi: 10.1371/journal.ppat.1007343. eCollection 2018 Oct. PLoS Pathog. 2018. PMID: 30346996 Free PMC article.
-
Prediction and molecular field view of drug resistance in HIV-1 protease mutants.Sci Rep. 2022 Feb 21;12(1):2913. doi: 10.1038/s41598-022-07012-x. Sci Rep. 2022. PMID: 35190671 Free PMC article.
-
Mechanistic insights into the substrate recognition of PPO: toward the rational design of effective inhibitors.Future Med Chem. 2014 Apr;6(6):597-9. doi: 10.4155/fmc.14.29. Future Med Chem. 2014. PMID: 24895889 Free PMC article.
-
Computational and experimental insights into the mechanism of substrate recognition and feedback inhibition of protoporphyrinogen oxidase.PLoS One. 2013 Jul 23;8(7):e69198. doi: 10.1371/journal.pone.0069198. Print 2013. PLoS One. 2013. PMID: 23935953 Free PMC article.
-
Free energy perturbation simulation on transition states and high-activity mutants of human butyrylcholinesterase for (-)-cocaine hydrolysis.J Phys Chem B. 2010 Aug 26;114(33):10889-96. doi: 10.1021/jp104989b. J Phys Chem B. 2010. PMID: 20677742 Free PMC article.
References
-
- Chaix-Couturier C, Holtzer C, Phillips KA, Durand-Zaleski I, Stansell J. Pharmacoecon. 2000;18:425–433. - PubMed
-
- Nitschko H, Gelderblom HR, Schatzl H, Von-der-Helm K. Int. Conf. AIDS. 1991;7:16–21.
-
- Supuran CT, Innocenti A, Mastrolorenzo A, Scozzafava A. Mini-Rev. Med. Chem. 2004;4:189–200. - PubMed
-
- Mastrolorenzo A, Rusconi S, Scozzafava A, Supuran CT. Expert Opin. Ther. Pat. 2006;16:1067–1091.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

