Morphological and molecular characterization of developing vertebral fusions using a teleost model
- PMID: 20604916
- PMCID: PMC2909226
- DOI: 10.1186/1472-6793-10-13
Morphological and molecular characterization of developing vertebral fusions using a teleost model
Abstract
Background: Spinal disorders are a major cause of disability for humans and an important health problem for intensively farmed animals. Experiments have shown that vertebral deformities present a complex but comparable etiology across species. However, the underlying molecular mechanisms involved in bone deformities are still far from understood. To further explicate the mechanisms involved, we have examined the fundamental aspects of bone metabolism and pathogenesis of vertebral fusions in Atlantic salmon (Salmo salar).
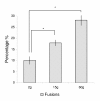
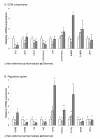
Results: Experimentally, juvenile salmon were subjected to hyperthermic conditions where more than 28% developed fused vertebral bodies. To characterize the fusion process we analyzed an intermediate and a terminal stage of the pathology by using x-ray, histology, immunohistochemistry, real-time quantitative PCR and in situ hybridization. At early stage in the fusion process, disorganized and proliferating osteoblasts were prominent at the growth zones of the vertebral body endplates. PCNA positive cells further extended along the rims of fusing vertebral bodies. During the developing pathology, the marked border between the osteoblast growth zones and the chondrocytic areas connected to the arches became less distinct, as proliferating cells and chondrocytes blended through an intermediate zone. This cell proliferation appeared to be closely linked to fusion of opposing arch centra. During the fusion process a metaplastic shift appeared in the arch centra where cells in the intermediate zone between osteoblasts and chondrocytes co-expressed mixed signals of chondrogenic and osteogenic markers. A similar shift also occurred in the notochord where proliferating chordoblasts changed transcription profile from chondrogenic to also include osteogenic marker genes. In progressed fusions, arch centra and intervertebral space mineralized.
Conclusion: Loss of cell integrity through cell proliferation and metaplastic shifts seem to be key events in the fusion process. The fusion process involves molecular regulation and cellular changes similar to those found in mammalian deformities, indicating that salmon is suitable for studying general bone development and to be a comparative model for spinal deformities.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

