Residues in the membrane-spanning domain core modulate conformation and fusogenicity of the HIV-1 envelope glycoprotein
- PMID: 20605619
- PMCID: PMC2902644
- DOI: 10.1016/j.virol.2010.03.016
Residues in the membrane-spanning domain core modulate conformation and fusogenicity of the HIV-1 envelope glycoprotein
Abstract
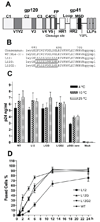
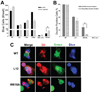
The membrane-spanning domain (MSD) of human immunodeficiency virus type I (HIV-1) envelope glycoprotein (Env) is critical for its biological activity. Initial studies have defined an almost invariant "core" structure in the MSD and demonstrated that it is crucial for anchoring Env in the membrane and virus entry. We show here that amino acid substitutions in the MSD "core" do not influence specific virus-cell attachment, nor CD4 receptor and CXCR4 coreceptor recognition by Env. However, substitutions within the MSD "core" delayed the kinetics and reduced the efficiency of cell-cell fusion mediated by Env. Although we observed no evidence that membrane fusion mediated by the MSD core mutants was arrested at a hemifusion stage, impaired Env fusogenicity was correlated with minor conformational changes in the V2, C1, and C5 regions in gp120 and the immunodominant loop in gp41. These changes could delay initiation of the conformational changes required in the fusion process.
2010 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection.J Virol. 2008 Jun;82(11):5417-28. doi: 10.1128/JVI.02666-07. Epub 2008 Mar 19. J Virol. 2008. PMID: 18353944 Free PMC article.
-
Characterization of Human Immunodeficiency Virus (HIV-1) Envelope Glycoprotein Variants Selected for Resistance to a CD4-Mimetic Compound.J Virol. 2022 Sep 14;96(17):e0063622. doi: 10.1128/jvi.00636-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980207 Free PMC article.
-
HIV-1 gp41 Residues Modulate CD4-Induced Conformational Changes in the Envelope Glycoprotein and Evolution of a Relaxed Conformation of gp120.J Virol. 2018 Jul 31;92(16):e00583-18. doi: 10.1128/JVI.00583-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875245 Free PMC article.
-
HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation.J Mol Biol. 2011 Jul 22;410(4):582-608. doi: 10.1016/j.jmb.2011.04.042. J Mol Biol. 2011. PMID: 21762802 Free PMC article. Review.
-
HIV-1 Envelope Conformation, Allostery, and Dynamics.Viruses. 2021 May 7;13(5):852. doi: 10.3390/v13050852. Viruses. 2021. PMID: 34067073 Free PMC article. Review.
Cited by
-
Beyond anchoring: the expanding role of the hendra virus fusion protein transmembrane domain in protein folding, stability, and function.J Virol. 2012 Mar;86(6):3003-13. doi: 10.1128/JVI.05762-11. Epub 2012 Jan 11. J Virol. 2012. PMID: 22238302 Free PMC article.
-
Functional bottlenecks for generation of HIV-1 intersubtype Env recombinants.Retrovirology. 2015 May 23;12:44. doi: 10.1186/s12977-015-0170-8. Retrovirology. 2015. PMID: 25997955 Free PMC article.
-
Characterization of the water defect at the HIV-1 gp41 membrane spanning domain in bilayers with and without cholesterol using molecular simulations.Biochim Biophys Acta. 2014 May;1838(5):1396-405. doi: 10.1016/j.bbamem.2014.01.009. Epub 2014 Jan 16. Biochim Biophys Acta. 2014. PMID: 24440660 Free PMC article.
-
HIV-1 Entry and Membrane Fusion Inhibitors.Viruses. 2021 Apr 23;13(5):735. doi: 10.3390/v13050735. Viruses. 2021. PMID: 33922579 Free PMC article. Review.
-
HIV-1 Envelope Glycoprotein Amino Acids Signatures Associated with Clade B Transmitted/Founder and Recent Viruses.Viruses. 2019 Nov 1;11(11):1012. doi: 10.3390/v11111012. Viruses. 2019. PMID: 31683782 Free PMC article.
References
-
- Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10(4):371–381. - PubMed
-
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. - PMC - PubMed
-
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–1027. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

