Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality
- PMID: 20606008
- PMCID: PMC2937546
- DOI: 10.1128/MCB.00218-10
Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality
Abstract
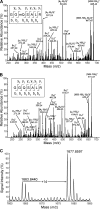
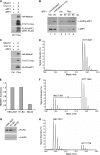
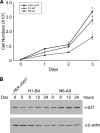
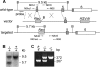
Biological methylation is a fundamental enzymatic reaction for a variety of substrates in multiple cellular processes. Mammalian N6amt1 was thought to be a homologue of bacterial N(6)-adenine DNA methyltransferases, but its substrate specificity and physiological importance remain elusive. Here, we demonstrate that N6amt1 functions as a protein methyltransferase for the translation termination factor eRF1 in mammalian cells both in vitro and in vivo. Mass spectrometry analysis indicated that about 70% of the endogenous eRF1 is methylated at the glutamine residue of the conserved GGQ motif. To address the physiological significance of eRF1 methylation, we disrupted the N6amt1 gene in the mouse. Loss of N6amt1 led to early embryonic lethality. The postimplantation development of mutant embryos was impaired, resulting in degeneration around embryonic day 6.5. This is in contrast to what occurs in Escherichia coli and Saccharomyces cerevisiae, which can survive without the N6amt1 homologues. Thus, N6amt1 is the first glutamine-specific protein methyltransferase characterized in vivo in mammals and methylation of eRF1 by N6amt1 might be essential for the viability of early embryos.
Figures


References
-
- Antonarakis, S. E., R. Lyle, S. Deutsch, and A. Reymond. 2002. Chromosome 21: a small land of fascinating disorders with unknown pathophysiology. Int. J. Dev. Biol. 46:89-96. - PubMed
-
- Bujnicki, J. M., and M. Radlinska. 1999. Is the HemK family of putative S-adenosylmethionine-dependent methyltransferases a “missing” zeta subfamily of adenine methyltransferases? A hypothesis. IUBMB Life 48:247-249. - PubMed
-
- Clarke, S. 1993. Protein methylation. Curr. Opin. Cell Biol. 5:977-983. - PubMed
-
- Figaro, S., N. Scrima, R. H. Buckingham, and V. Heurgue-Hamard. 2008. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 582:2352-2356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
