Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study
- PMID: 20606121
- PMCID: PMC2910759
- DOI: 10.1161/CIRCULATIONAHA.109.928192
Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study
Abstract
Background: Although many genetic epidemiology and biomarker studies have been conducted to examine associations of genetic variants and circulating proteins with cardiovascular disease and risk factors, there has been little study of gene expression or transcriptomics. Quantitative differences in the abundance of transcripts has been demonstrated in malignancies, but gene expression from a large community-based cohort examining risk of cardiovascular disease has never been reported.
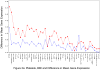
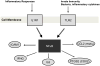
Methods and results: On the basis of preliminary microarray data and previously suggested genes from the literature, we measured expression of 48 genes by high-throughput quantitative reverse-transcriptase polymerase chain reaction in 1846 participants of the Framingham Offspring cohort from RNA derived from isolated platelets and leukocytes. A multivariable stepwise regression model was used to assess clinical correlates of quantitative RNA expression. For specific inflammatory platelet-derived transcripts, including ICAM1, IFNG, IL1R1, IL6, MPO, COX2, TNF, TLR2, and TLR4, there were significant associations with higher body mass index (BMI). Compared with platelets, fewer leukocyte-derived transcripts were associated with BMI or other cardiovascular risk factors. Select transcripts were found to be highly heritable, including GPIBA and COX1. Almost uniformly, heritable transcripts were not those associated with BMI.
Conclusions: Inflammatory transcripts derived from platelets, particularly those part of the nuclear factor kappa B pathway, are associated with BMI, whereas others are heritable. This is the first study, using a large community-based cohort, to demonstrate clinical correlates of gene expression and is consistent with the hypothesis that specific peripheral-blood transcripts play a role in the pathogenesis of coronary heart disease and its risk factors.
Conflict of interest statement
The authors have no conflicts of interest to report.
Figures


References
-
- Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, MacCluer JW, Kissebah AH, Collier GR, Moses EK, Blangero J. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. - PubMed
-
- Greiner TC. mRNA microarray analysis in lymphoma and leukemia. Cancer Treat Res. 2004;121:1–12. - PubMed
-
- Zent CS, Zhan F, Schichman SA, Bumm KH, Lin P, Chen JB, Shaughnessy JD. The distinct gene expression profiles of chronic lymphocytic leukemia and multiple myeloma suggest different anti-apoptotic mechanisms but predict only some differences in phenotype. Leuk Res. 2003;27:765–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

