Kinetic and spectroscopic evidence of negative cooperativity in the action of lysine 2,3-aminomutase
- PMID: 20608698
- PMCID: PMC4337230
- DOI: 10.1021/jp103856m
Kinetic and spectroscopic evidence of negative cooperativity in the action of lysine 2,3-aminomutase
Abstract
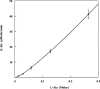
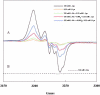
Lysine 2,3-aminomutase (LAM) catalyzes the interconversion of L-lysine and L-β-lysine, a component of a number of antibiotics. The reaction requires the cofactors S-adenosyl-L-methionine (SAM), pyridoxal-5'-phosphate (PLP), and a [4Fe-4S] cluster. LAM is a founding member of the radical SAM superfamily of enzymes. LAM is highly specific for L-lysine and will not accept most other amino acids as substrates. L-alanine and L-2-aminobutyrate at 0.2 M react as substrates for LAM at, respectively, 5 × 10(-6) and 8 × 10(-5) times the rate with saturating L-lysine. Saturating ethylamine accelerates the L-alanine reaction 70-fold, and saturating methylamine accelerates the L-2-aminobutyrate reaction 47-fold. The primary amines binding at the active site of LAM with L-alanine or L-2-aminobutyrate simulate L-lysine. The steady-state kinetics of the reaction of L-alanine + ethylamine displays negative cooperativity with respect to L-alanine. The second-order rate constant for production of β-alanine in the reaction of L-alanine and saturating ethylamine is 0.040 M(-1) s(-1), which is 2 × 10(-5) times the value of k(cat)/K(m) for the reaction of L-lysine. When L-lysine is at a concentration 1/16th of K(m), the lysyl-free radical intermediate is hardly detectable by EPR; however, the addition of L-alanine at high concentration (0.2 M) enhances free radical formation, and the addition of ethylamine further enhances radical formation. These facts complement the kinetic observations and support negative cooperativity in the reaction of L-alanine as a substrate for LAM. Present results and independent evidence support negative cooperativity in the reaction of L-lysine as well.
Figures

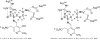
Similar articles
-
Large-scale domain motions and pyridoxal-5'-phosphate assisted radical catalysis in coenzyme B12-dependent aminomutases.Int J Mol Sci. 2014 Feb 20;15(2):3064-87. doi: 10.3390/ijms15023064. Int J Mol Sci. 2014. PMID: 24562332 Free PMC article. Review.
-
Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme.Biochemistry. 2007 Nov 13;46(45):12889-95. doi: 10.1021/bi701745h. Epub 2007 Oct 18. Biochemistry. 2007. PMID: 17944492 Free PMC article.
-
Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe-4S] center in lysine 2,3-aminomutase.J Am Chem Soc. 2003 Oct 1;125(39):11788-9. doi: 10.1021/ja036120z. J Am Chem Soc. 2003. PMID: 14505379
-
S-Adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance.Biochemistry. 1998 Feb 24;37(8):2578-85. doi: 10.1021/bi972417w. Biochemistry. 1998. PMID: 9485408
-
The role of radicals in enzymatic processes.Chem Rec. 2001;1(4):277-89. doi: 10.1002/tcr.1013. Chem Rec. 2001. PMID: 11893068 Review.
Cited by
-
Large-scale domain motions and pyridoxal-5'-phosphate assisted radical catalysis in coenzyme B12-dependent aminomutases.Int J Mol Sci. 2014 Feb 20;15(2):3064-87. doi: 10.3390/ijms15023064. Int J Mol Sci. 2014. PMID: 24562332 Free PMC article. Review.
-
Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus: Biochemical and Structural Characterization.Front Bioeng Biotechnol. 2019 Jan 24;7:7. doi: 10.3389/fbioe.2019.00007. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 30733943 Free PMC article.
-
Cooperativity in Binding Processes: New Insights from Phenomenological Modeling.PLoS One. 2015 Dec 30;10(12):e0146043. doi: 10.1371/journal.pone.0146043. eCollection 2015. PLoS One. 2015. PMID: 26717487 Free PMC article.
-
Mechanistic Studies of Radical SAM Enzymes: Pyruvate Formate-Lyase Activating Enzyme and Lysine 2,3-Aminomutase Case Studies.Methods Enzymol. 2018;606:269-318. doi: 10.1016/bs.mie.2018.04.013. Epub 2018 Jul 7. Methods Enzymol. 2018. PMID: 30097096 Free PMC article.
References
-
- Frey P,A, Hegeman AD, Ruzicka FJ. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. - PubMed
-
- Frey PA, Th O. Magnusson, Chem. Rev. 2003;103:2129–2148. - PubMed
-
- Ballinger MD, Reed GH, Frey PA. Biochemistry. 1992;31:949–953. - PubMed
-
- Ballinger MD, Frey PA, Reed GH. Biochemistry. 1992;31:10782–10789. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

