Why genes overlap in viruses
- PMID: 20610432
- PMCID: PMC2992710
- DOI: 10.1098/rspb.2010.1052
Why genes overlap in viruses
Abstract
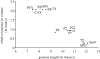
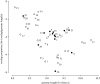
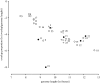
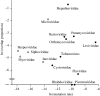
The genomes of most virus species have overlapping genes--two or more proteins coded for by the same nucleotide sequence. Several explanations have been proposed for the evolution of this phenomenon, and we test these by comparing the amount of gene overlap in all known virus species. We conclude that gene overlap is unlikely to have evolved as a way of compressing the genome in response to the harmful effect of mutation because RNA viruses, despite having generally higher mutation rates, have less gene overlap on average than DNA viruses of comparable genome length. However, we do find a negative relationship between overlap proportion and genome length among viruses with icosahedral capsids, but not among those with other capsid types that we consider easier to enlarge in size. Our interpretation is that a physical constraint on genome length by the capsid has led to gene overlap evolving as a mechanism for producing more proteins from the same genome length. We consider that these patterns cannot be explained by other factors, namely the possible roles of overlap in transcription regulation, generating more divergent proteins and the relationship between gene length and genome length.
Figures
References
-
- Agranovsky A. A.1996The principles of molecular organization, expression and evolution of closteroviruses: over the barriers. Adv. Virus Res. 47, 119–15810.1016/S0065-3527(08)60735-6 (doi:10.1016/S0065-3527(08)60735-6) - DOI - DOI - PMC - PubMed
-
- Ahlquist P.2005Virus evolution: fitting lifestyles to a T. Curr. Biol. 15, R465–R46710.1016/j.cub.2005.06.016 (doi:10.1016/j.cub.2005.06.016) - DOI - DOI - PubMed
-
- Bamford D. H., Grimes J. M., Stuart D. I.2005What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15, 655–66310.1016/j.sbi.2005.10.012 (doi:10.1016/j.sbi.2005.10.012) - DOI - DOI - PubMed
-
- Barrell B. G., Air G. M., Hutchison C. A.1976Overlapping genes in bacteriophage-φχ174. Nature 264, 34–4110.1038/264034a0 (doi:10.1038/264034a0) - DOI - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

