Role of peroxiredoxin 1 and peroxiredoxin 4 in protection of respiratory syncytial virus-induced cysteinyl oxidation of nuclear cytoskeletal proteins
- PMID: 20610706
- PMCID: PMC2937607
- DOI: 10.1128/JVI.01005-10
Role of peroxiredoxin 1 and peroxiredoxin 4 in protection of respiratory syncytial virus-induced cysteinyl oxidation of nuclear cytoskeletal proteins
Abstract
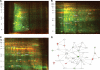
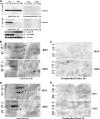
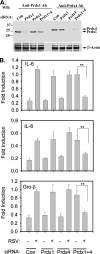
The respiratory epithelium plays a central role in innate immunity by secreting networks of inflammatory mediators in response to respiratory syncytial virus (RSV) infection. Previous proteomic studies focusing on the host cellular response to RSV indicated the existence of a nuclear heat shock response and cytoplasmic depletion of antioxidant proteins in model type II-like airway epithelial cells. Here, we increased the depth of nuclear proteomic interrogation by using fluorescence difference labeling followed by liquid isoelectric focusing prefractionation/two-dimensional gel electrophoresis (2-DE) to identify an additional 41 proteins affected by RSV infection. Surprisingly, we found inducible oligomers and shifts in isoelectric points for peroxiredoxin 1 (Prdx-1), Prdx-3, and Prdx-4 isoforms without changes in their total abundance, indicating that Prdxs were being oxidized in response to RSV. To address the role of Prdx-1 and Prdx-4 in RSV infection, isoforms were selectively knocked down by small interfering RNA (siRNA) transfection. Cells lacking Prdx-1, Prdx-4, or both showed increased levels of reactive oxygen species formation and a higher level of protein carbonylation in response to RSV infection. Using a novel saturation fluorescence labeling 2-DE analysis, we showed that 15 unique proteins had enhanced oxidative modifications of at least >1.2-fold in the Prdx knockdowns in response to RSV, including annexin A2 and desmoplakin. Our results suggest that Prdx-1 and Prdx-4 are essential for preventing RSV-induced oxidative damage in a subset of nuclear intermediate filament and actin binding proteins in epithelial cells.
Figures

References
-
- Brasier, A. R., H. Spratt, Z. Wu, I. Boldogh, Y. Zhang, R. P. Garofalo, A. Casola, J. Pashmi, A. Haag, B. Luxon, and A. Kurosky. 2004. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected A549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78:11461-11476. - PMC - PubMed
-
- Caplan, J. F., N. R. Filipenko, S. L. Fitzpatrick, and D. M. Waisman. 2004. Regulation of annexin A2 by reversible glutathionylation. J. Biol. Chem. 279:7740-7750. - PubMed
-
- Casola, A., N. Burger, T. Liu, M. Jamaluddin, A. R. Brasier, and R. P. Garofalo. 2001. Oxidant tone regulates RANTES gene expression in airway epithelial cells infected with respiratory syncytial virus. Role in viral-induced interferon regulatory factor activation. J. Biol. Chem. 276:19715-19722. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

