The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule?
- PMID: 20613982
- PMCID: PMC2894941
- DOI: 10.1371/journal.pone.0011387
The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule?
Abstract
Background: During typical microRNA (miRNA) biogenesis, one strand of a approximately 22 nt RNA duplex is preferentially selected for entry into a silencing complex, whereas the other strand, known as the passenger strand or miRNA* strand, is degraded. Recently, some miRNA* sequences were reported as guide miRNAs with abundant expression. Here, we intended to discover evolutionary implication of the fate of miRNA* strand by analyzing miRNA/miRNA* sequences across vertebrates.
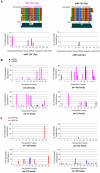
Principal findings: Mature miRNAs based on gene families were well conserved especially for their seed sequences across vertebrates, while their passenger strands always showed various divergence patterns. The divergence mainly resulted from divergence of different animal species, homologous miRNA genes and multicopy miRNA hairpin precursors. Some miRNA* sequences were phylogenetically conserved in seed and anchor sequences similar to mature miRNAs, while others revealed high levels of nucleotide divergence despite some of their partners were highly conserved. Most of those miRNA precursors that could generate abundant miRNAs from both strands always were well conserved in sequences of miR-#-5p and miR-#-3p, especially for their seed sequences.
Conclusions: The final fate of miRNA* strand, either degraded as merely carrier strand or expressed abundantly as potential functional guide miRNA, may be destined across evolution. Well-conserved miRNA* strands, particularly conservation in seed sequences, maybe afford potential opportunities for contributing to regulation network. The study will broaden our understanding of potential functional miRNA* species.
Conflict of interest statement
Figures




References
-
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Plasterk RHA. Micro RNAs in animal development. Cell. 2006;124:877–881. - PubMed
-
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature Structural & Molecular Biology. 2006;13:1097–1101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

