Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function
- PMID: 20614009
- PMCID: PMC2894074
- DOI: 10.1371/journal.pone.0011379
Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function
Abstract
Background: Approximately half of hereditary breast cancers have mutations in either BRCA1 or BRCA2. BRCA1 is a multifaceted tumor suppressor protein that has implications in processes such as cell cycle, transcription, DNA damage response and chromatin remodeling. This multifunctional nature of BRCA1 is achieved by exerting its many effects through modulation of transcription. Many cellular events are dictated by covalent modification of proteins, an important mechanism in regulating protein and genome function; of which protein methylation is an important posttranslational modification with activating or repressive effects.
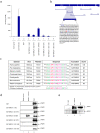
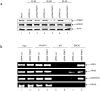
Methods/principal findings: Here we demonstrate for the first time that BRCA1 is methylated both in breast cancer cell lines and breast cancer tumor samples at arginine and lysine residues through immunoprecipitation and western blot analysis. Arginine methylation by PRMT1 was observed in vitro and the region of BRCA1 504-802 shown to be highly methylated. PRMT1 was detected in complex with BRCA1 504-802 through in vitro binding assays and co-immunoprecipitated with BRCA1. Inhibition of methylation resulted in decreased BRCA1 methylation and alteration of BRCA1 binding to promoters in vivo as shown through chromatin immunoprecipitation assays. Knockdown of PRMT1 also resulted in increased BRCA1 binding to particular promoters in vivo. Finally, following methylation inhibition, Sp1 was found to preferentially associate with hypo-methylated BRCA1 and STAT1 was found to preferentially associate with hyper-methylated BRCA1.
Conclusions/significance: These results suggest that methylation may influence either the ability of BRCA1 to bind to specific promoters or protein-protein interactions which alters the recruitment of BRCA1 to these promoters. Thus, given the importance of BRCA1 to genomic stability, methylation of BRCA1 may ultimately affect the tumor suppressor ability of BRCA1.
Conflict of interest statement
Figures




References
-
- Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579–585. - PubMed
-
- Krum SA, Miranda GA, Lin C, Lane TF. BRCA1 associates with processive RNA polymerase II. J Biol Chem. 2003;278:52012–52020. - PubMed
-
- Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat Genet. 1998;19:254–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

