Iron-nucleated folding of a metalloprotein in high urea: resolution of metal binding and protein folding events
- PMID: 20614892
- PMCID: PMC2914805
- DOI: 10.1021/bi100630t
Iron-nucleated folding of a metalloprotein in high urea: resolution of metal binding and protein folding events
Abstract
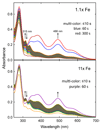
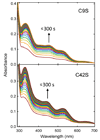
Addition of iron salts to chaotrope-denatured aporubredoxin (apoRd) leads to nearly quantitative recovery of its single Fe(SCys)(4) site and native protein structure without significant dilution of the chaotrope. This "high-chaotrope" approach was used to examine iron binding and protein folding events using stopped-flow UV-vis absorption and CD spectroscopies. With a 100-fold molar excess of ferrous iron over denatured apoRd maintained in 5 M urea, the folded holoFe(III)Rd structure was recovered in >90% yield with a t(1/2) of <10 ms. More modest excesses of iron also gave nearly quantitative holoRd formation in 5 M urea but with chronological resolution of iron binding and protein folding events. The results indicate structural recovery in 5 M urea consists of the minimal sequence: (1) binding of ferrous iron to the unfolded apoRd, (2) rapid formation of a near-native ferrous Fe(SCys)(4) site within a protein having no detectable secondary structure, and (3) recovery of the ferrous Fe(SCys)(4) site chiral environment nearly concomitantly with (4) recovery of the native protein secondary structure. The rate of step 2 (and, by inference, step 1) was not saturated even at a 100-fold molar excess of iron. Analogous results obtained for Cys --> Ser iron ligand variants support formation of an unfolded-Fe(SCys)(3) complex between steps 1 and 2, which we propose is the key nucleation event that pulls together distal regions of the protein chain. These results show that folding of chaotrope-denatured apoRd is iron-nucleated and driven by extraordinarily rapid formation of the Fe(SCys)(4) site from an essentially random coil apoprotein. This high-chaotrope, multispectroscopy approach could clarify folding pathways of other [M(SCys)(3)]- or [M(SCys)(4)]-containing proteins.
Figures




Similar articles
-
"Iron priming" guides folding of denatured aporubredoxins.J Biol Inorg Chem. 2008 Aug;13(6):981-91. doi: 10.1007/s00775-008-0385-4. Epub 2008 Apr 30. J Biol Inorg Chem. 2008. PMID: 18446387 Free PMC article.
-
Rubredoxin refolding on nanostructured hydrophobic surfaces: evidence for a new type of biomimetic chaperones.Proteins. 2014 Nov;82(11):3154-62. doi: 10.1002/prot.24675. Epub 2014 Sep 3. Proteins. 2014. PMID: 25143010
-
Contribution of the [FeII(SCys)4] site to the thermostability of rubredoxins.J Biol Inorg Chem. 2004 Apr;9(3):297-306. doi: 10.1007/s00775-004-0525-4. Epub 2004 Feb 10. J Biol Inorg Chem. 2004. PMID: 14770302
-
Role of cofactors in metalloprotein folding.Q Rev Biophys. 2004 Aug-Nov;37(3-4):285-314. doi: 10.1017/S003358350500404X. Q Rev Biophys. 2004. PMID: 16194296 Review.
-
Residual ordered structure in denatured proteins and the problem of protein folding.Indian J Biochem Biophys. 2012 Feb;49(1):7-17. Indian J Biochem Biophys. 2012. PMID: 22435139 Review.
Cited by
-
Treponema denticola superoxide reductase: in vivo role, in vitro reactivities, and a novel [Fe(Cys)(4)] site.Biochemistry. 2012 Jul 17;51(28):5601-10. doi: 10.1021/bi300667s. Epub 2012 Jun 29. Biochemistry. 2012. PMID: 22715932 Free PMC article.
-
Direct measurements of the mechanical stability of zinc-thiolate bonds in rubredoxin by single-molecule atomic force microscopy.Biophys J. 2011 Sep 21;101(6):1467-73. doi: 10.1016/j.bpj.2011.08.021. Epub 2011 Sep 20. Biophys J. 2011. PMID: 21943428 Free PMC article.
-
Metal selectivity of the Escherichia coli nickel metallochaperone, SlyD.Biochemistry. 2011 Dec 13;50(49):10666-77. doi: 10.1021/bi2014882. Epub 2011 Nov 14. Biochemistry. 2011. PMID: 22047179 Free PMC article.
-
Cluster and fold stability of E. coli ISC-type ferredoxin.PLoS One. 2013 Nov 12;8(11):e78948. doi: 10.1371/journal.pone.0078948. eCollection 2013. PLoS One. 2013. PMID: 24265733 Free PMC article.
-
Protein interactions in the biological assembly of iron-sulfur clusters in Escherichia coli: Molecular and mechanistic aspects of the earliest assembly steps.IUBMB Life. 2022 Jul;74(7):723-732. doi: 10.1002/iub.2622. Epub 2022 May 25. IUBMB Life. 2022. PMID: 35611886 Free PMC article.
References
-
- Cavagnero S, Zhou ZH, Adams MW, Chan SI. Unfolding mechanism of rubredoxin from Pyrococcus furiosus. Biochemistry. 1998;37:3377–3385. - PubMed
-
- LeMaster DM, Hernandez G. Additivity in both thermodynamic stability and thermal transition temperature for rubredoxin chimeras via hybrid native partitioning. Structure. 2005;13:1153–1163. - PubMed
-
- Henriques BJ, Saraiva LM, Gomes CM. Combined spectroscopic and calorimetric characterisation of rubredoxin reversible thermal transition. J. Biol. Inorg. Chem. 2006;11:73–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

