What are nuclear receptor ligands?
- PMID: 20615454
- PMCID: PMC3010294
- DOI: 10.1016/j.mce.2010.06.018
What are nuclear receptor ligands?
Abstract
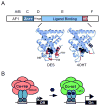
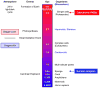
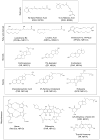
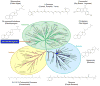
Nuclear receptors (NRs) are a family of highly conserved transcription factors that regulate transcription in response to small lipophilic compounds. They play a role in every aspect of development, physiology and disease in humans. They are also ubiquitous in and unique to the animal kingdom suggesting that they may have played an important role in their evolution. In contrast to the classical endocrine receptors that originally defined the family, recent studies suggest that the first NRs might have been sensors of their environment, binding ligands that were external to the host organism. The purpose of this review is to provide a broad perspective on NR ligands and address the issue of exactly what constitutes a NR ligand from historical, biological and evolutionary perspectives. This discussion will lay the foundation for subsequent reviews in this issue as well as pose new questions for future investigation.
Copyright © 2010 Elsevier Ireland Ltd. All rights reserved.
Figures

References
-
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. - PubMed
-
- Al-Moghrabi S, Allemand D, Couret JM, Jaubert J. Fatty acids of the scleractinian coral Galaxea fascicularis: effect of light and feeding. J Comp Physiol B. 1995;165:183–192.
-
- Baker ME. Trichoplax, the simplest known animal, contains an estrogen-related receptor but no estrogen receptor: Implications for estrogen receptor evolution. Biochem Biophys Res Commun. 2008;375:623–627. - PubMed
-
- Baldauf SL, Bhattacharya D, Cockrill J, Hugenholtz P, Pawlowski J, Simpson AGB. In: The Tree of Life: A Overview. Craycraft J, Donoghue MJ, editors. Assembling the Tree of Life Oxford University Press; 2004. pp. 43–75.
-
- Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14:369–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

