Export of microRNAs and microRNA-protective protein by mammalian cells
- PMID: 20615901
- PMCID: PMC2978372
- DOI: 10.1093/nar/gkq601
Export of microRNAs and microRNA-protective protein by mammalian cells
Abstract
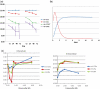
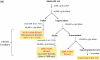
The discovery of microRNAs (miRNAs) as a new class of regulators of gene expression has triggered an explosion of research activities, but has left many unanswered questions about how this regulation functions and how it is integrated with other regulatory mechanisms. A number of miRNAs have been found to be present in plasma and other body fluids of humans and mice in surprisingly high concentrations. This observation was unexpected in two respects: first, the fact that these molecules are present at all outside the cell at significant concentrations and second, that these molecules appear to be stable outside of the cell. In light of this it has been suggested that the biological function of miRNAs may also extend outside of the cell and mediate cell-cell communication. We report here that after serum deprivation several human cell lines tested promptly export a substantial amount of miRNAs into the culture medium and the export process is largely energy dependent. The exported miRNAs are found both within and outside of the 16.5 and 120 K centrifugation pellets which contain most of the known cell-derived vesicles, the microvesicles and exosomes. We have identified some candidate proteins involved in this system, and one of these proteins may also play a role in protecting extracellular miRNAs from degradation. Our results point to a hitherto unrecognized and uncharacterized miRNA trafficking system in mammalian cells that is consistent with the cell-cell communication hypothesis.
Figures





References
-
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. - PubMed
-
- Omer AD, Janas MM, Novina CD. The chicken or the egg: microRNA-mediated regulation of mRNA translation or mRNA stability. Mol. Cell. 2009;35:739–740. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

