Global analysis of trans-splicing in Drosophila
- PMID: 20615941
- PMCID: PMC2919919
- DOI: 10.1073/pnas.1007586107
Global analysis of trans-splicing in Drosophila
Erratum in
- Proc Natl Acad Sci U S A. 2013 May 7;110(19):7958
Abstract
Precursor mRNA (pre-mRNA) splicing can join exons contained on either a single pre-mRNA (cis) or on separate pre-mRNAs (trans). It is exceedingly rare to have trans-splicing between protein-coding exons and has been demonstrated for only two Drosophila genes: mod(mdg4) and lola. It has also been suggested that trans-splicing is a mechanism for the generation of chimeric RNA products containing sequence from multiple distant genomic sites. Because most high-throughput approaches cannot distinguish cis- and trans-splicing events, the extent to which trans-splicing occurs between protein-coding exons in any organism is unknown. Here, we used paired-end deep sequencing of mRNA to identify genes that undergo trans-splicing in Drosophila interspecies hybrids. We did not observe credible evidence for the existence of chimeric RNAs generated by trans-splicing of RNAs transcribed from distant genomic loci. Rather, our data suggest that experimental artifacts are the source of most, if not all, apparent chimeric RNA products. We did, however, identify 80 genes that appear to undergo trans-splicing between homologous alleles and can be classified into three categories based on their organization: (i) genes with multiple 3' terminal exons, (ii) genes with multiple first exons, and (iii) genes with very large introns, often containing other genes. Our results suggest that trans-splicing between homologous alleles occurs more commonly in Drosophila than previously believed and may facilitate expression of architecturally complex genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
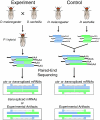

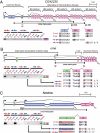
References
-
- Konarska MM, Padgett RA, Sharp PA. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. - PubMed
-
- Solnick D. Trans splicing of mRNA precursors. Cell. 1985;42:157–164. - PubMed
-
- Nilsen TW. Evolutionary origin of SL-addition trans-splicing: Still an enigma. Trends Genet. 2001;17:678–680. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

