A time-invariant principle of genome evolution
- PMID: 20615949
- PMCID: PMC2919972
- DOI: 10.1073/pnas.0914454107
A time-invariant principle of genome evolution
Abstract
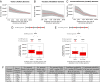
Uncovering general principles of genome evolution that are time-invariant and that operate in germ and somatic cells has implications for genome-wide association studies (GWAS), gene therapy, and disease genomics. Here we investigate the relationship between structural alterations (e.g., insertions and deletions) and single-nucleotide substitutions by comparing the following genomes that diverged at different times across germ- and somatic-cell lineages: (i) the reference human and chimpanzee genome (in million years), (ii) the reference human and personal genomes (in tens of thousands of years), and (iii) structurally altered regions in cancer and genetically engineered cells (in days). At the species level, genes with structural alteration in nearby regions show increased single-nucleotide changes and tend to evolve faster. In personal genomes, the single-nucleotide substitution rate is higher near sites of structural alteration and decreases with increasing distance. In human cancer cell populations and in cells genetically engineered using zinc-finger nucleases, single-nucleotide changes occur frequently near sites of structural alterations. We present evidence that structural alteration induces single-nucleotide changes in nearby regions and discuss possible molecular mechanisms that contribute to this phenomenon. We propose that the low fidelity of nonreplicative error-prone repair polymerases, which are used during insertion or deletion, result in break-repair-induced single-nucleotide mutations in the vicinity of structural alteration. Thus, in the mutational landscape, structural alterations are linked to single-nucleotide changes across different time scales in both somatic- and germ-cell lineages. We discuss implications for genome evolution, GWAS, disease genomics, and gene therapy and emphasize the need to investigate both types of mutations within a single framework.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Mutation rate: DNA repair and indels boost errors.Nat Rev Genet. 2010 Sep;11(9):592. doi: 10.1038/nrg2848. Epub 2010 Jul 27. Nat Rev Genet. 2010. PMID: 20661254 No abstract available.
References
-
- Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press, Oxford; 1930.
-
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence: Accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. - PubMed
-
- Tian D, et al. Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature. 2008;455:105–108. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

