Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex
- PMID: 20615956
- PMCID: PMC2919950
- DOI: 10.1073/pnas.1002285107
Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex
Abstract
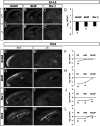
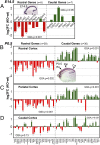
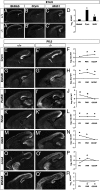
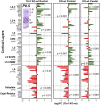
Areas and layers of the cerebral cortex are specified by genetic programs that are initiated in progenitor cells and then, implemented in postmitotic neurons. Here, we report that Tbr1, a transcription factor expressed in postmitotic projection neurons, exerts positive and negative control over both regional (areal) and laminar identity. Tbr1 null mice exhibited profound defects of frontal cortex and layer 6 differentiation, as indicated by down-regulation of gene-expression markers such as Bcl6 and Cdh9. Conversely, genes that implement caudal cortex and layer 5 identity, such as Bhlhb5 and Fezf2, were up-regulated in Tbr1 mutants. Tbr1 implements frontal identity in part by direct promoter binding and activation of Auts2, a frontal cortex gene implicated in autism. Tbr1 regulates laminar identity in part by downstream activation or maintenance of Sox5, an important transcription factor controlling neuronal migration and corticofugal axon projections. Similar to Sox5 mutants, Tbr1 mutants exhibit ectopic axon projections to the hypothalamus and cerebral peduncle. Together, our findings show that Tbr1 coordinately regulates regional and laminar identity of postmitotic cortical neurons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

