Microtubule-assisted mechanism for functional metabolic macromolecular complex formation
- PMID: 20615962
- PMCID: PMC2919939
- DOI: 10.1073/pnas.1008451107
Microtubule-assisted mechanism for functional metabolic macromolecular complex formation
Abstract
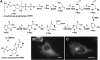
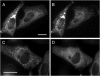
Evidence has been presented for a metabolic multienzyme complex, the purinosome, that participates in de novo purine biosynthesis to form clusters in the cytoplasm of living cells under purine-depleted conditions. Here we identified, using fluorescent live cell imaging, that a microtubule network appears to physically control the spatial distribution of purinosomes in the cytoplasm. Application of a cell-based assay measuring the rate of de novo purine biosynthesis confirmed that the metabolic activity of purinosomes was significantly suppressed in the absence of microtubules. Collectively, we propose a microtubule-assisted mechanism for functional purinosome formation in HeLa cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rowe PB, Wyngaarden JB. Glutamine phosphoribosylpyrophosphate amidotransferase. Purification, substructure, amino acid composition, and absorption spectra. J Biol Chem. 1968;243:6373–6383. - PubMed
-
- Smith GK, Mueller WT, Wasserman GF, Taylor WD, Benkovic SJ. Characterization of the enzyme complex involving the folate-requiring enzymes of de novo purine biosynthesis. Biochemistry. 1980;19:4313–4321. - PubMed
-
- Srere PA. Complexes of sequential metabolic enzymes. Annu Rev Biochem. 1987;56:89–124. - PubMed
-
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320:103–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

