RNA-mediated trans-communication can establish paramutation at the b1 locus in maize
- PMID: 20616013
- PMCID: PMC2919911
- DOI: 10.1073/pnas.1007972107
RNA-mediated trans-communication can establish paramutation at the b1 locus in maize
Abstract
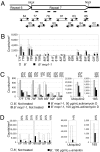
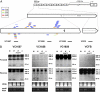
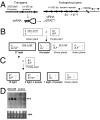
Paramutation is the epigenetic transfer of information between alleles that leads to the heritable change of expression of one allele. Paramutation at the b1 locus in maize requires seven noncoding tandem repeat (b1TR) sequences located approximately 100 kb upstream of the transcription start site of b1, and mutations in several genes required for paramutation implicate an RNA-mediated mechanism. The mediator of paramutation (mop1) gene, which encodes a protein closely related to RNA-dependent RNA polymerases, is absolutely required for paramutation. Herein, we investigate the potential function of mop1 and the siRNAs that are produced from the b1TR sequences. Production of siRNAs from the b1TR sequences depends on a functional mop1 gene, but transcription of the repeats is not dependent on mop1. Further nuclear transcription assays suggest that the b1TR sequences are likely transcribed predominantly by RNA polymerase II. To address whether production of b1TR-siRNAs correlated with paramutation, we examined siRNA production in alleles that cannot undergo paramutation. Alleles that cannot participate in paramutation also produce b1TR-siRNAs, suggesting that b1TR-siRNAs are not sufficient for paramutation in the tissues analyzed. However, when b1TR-siRNAs are produced from a transgene expressing a hairpin RNA, b1 paramutation can be recapitulated. We hypothesize that either the b1TR-siRNAs or the dsRNA template mediates the trans-communication between the alleles that establishes paramutation. In addition, we uncovered a role for mop1 in the biogenesis of a subset of microRNAs (miRNAs) and show that it functions at the level of production of the primary miRNA transcripts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

