Role of Kruppel-like factors in leukocyte development, function, and disease
- PMID: 20616217
- PMCID: PMC2996110
- DOI: 10.1182/blood-2010-05-285353
Role of Kruppel-like factors in leukocyte development, function, and disease
Abstract
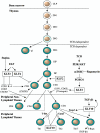
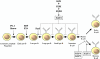
The Krüppel-like transcription factor (KLF) family participates in diverse aspects of cellular growth, development, differentiation, and activation. Recently, several groups have identified new connections between the function of these factors and leukocyte responses in health and disease. Gene targeting of individual KLFs in mice has uncovered novel and unexpected physiologic roles among myeloid and lymphocyte cell lineage maturation, particularly in the bone marrow niche and blood. In addition, several KLF family members are downstream targets of stimuli and signaling pathways critical to T-cell trafficking, T regulatory cell differentiation or suppressor function, monocyte/macrophage activation or renewal, and B memory cell maturation or activation. Indeed, KLFs have been implicated in subtypes of leukemia, lymphoma, autoimmunity, and in acute and chronic inflammatory disease states, such as atherosclerosis, diabetes, and airway inflammation, raising the possibility that KLFs and their upstream signals are of therapeutic interest. This review focuses on the relevant literature of Krüppel-like factors in leukocyte biology and their implications in clinical settings.
Figures


References
-
- Wieschaus E, Nusslein-Volhard C, Kluding H. Kruppel, a gene whose activity is required early in the zygotic genome for normal embryonic segmentation. Dev Biol. 1984;104(1):172–186. - PubMed
-
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. - PubMed
-
- Preiss A, Rosenberg UB, Kienlin A, Seifert E, Jackle H. Molecular genetics of Kruppel, a gene required for segmentation of the Drosophila embryo. Nature. 1985;313(5997):27–32. - PubMed
-
- Jackle H, Rosenberg UB, Preiss A, et al. Molecular analysis of Kruppel, a segmentation gene of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:465–473. - PubMed
-
- Schuh R, Aicher W, Gaul U, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell. 1986;47(6):1025–1032. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

