Human amebiasis: breaking the paradigm?
- PMID: 20617021
- PMCID: PMC2872301
- DOI: 10.3390/ijerph7031105
Human amebiasis: breaking the paradigm?
Abstract
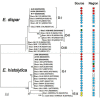
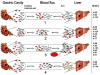
For over 30 years it has been established that the Entamoeba histolytica protozoan included two biologically and genetically different species, one with a pathogenic phenotype called E. histolytica and the other with a non-pathogenic phenotype called Entamoeba dispar. Both of these amoebae species can infect humans. E. histolytica has been considered as a potential pathogen that can cause serious damage to the large intestine (colitis, dysentery) and other extraintestinal organs, mainly the liver (amebic liver abscess), whereas E. dispar is a species that interacts with humans in a commensal relationship, causing no symptoms or any tissue damage. This paradigm, however, should be reconsidered or re-evaluated. In the present work, we report the detection and genotyping of E. dispar sequences of DNA obtained from patients with amebic liver abscesses, including the genotyping of an isolate obtained from a Brazilian patient with a clinical diagnosis of intestinal amebiasis that was previously characterized as an E. dispar species. The genetic diversity and phylogenetic analysis performed by our group has shown the existence of several different genotypes of E. dispar that can be associated to, or be potentiality responsible for intestinal or liver tissue damage, similar to that observed with E. histolytica.
Keywords: E. dispar; E. histolytica; genetic diversity; human amebiasis; phylogeny.
Figures

References
-
- Sargeaunt PG, Jackson TFHG, Wiffen SR, Bhojnani R. Bological evidence of genetic exchange in Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 1988;82:862–867. - PubMed
-
- Sargeaunt PG. Zymodemes expressing possible genetic exchange in Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 1985;79:86–89. - PubMed
-
- World Health Organization Amoebiasis. Wkly. Epidemiol. Rec. 1997;72:97–100. - PubMed
-
- Walsh JA. Problems in recognition and diagnosis of amebiasis: estimation of the global magnitude of the morbidity and mortality. Rev. Infect. Dis. 1986;8:228–238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

