The landscape genetics of infectious disease emergence and spread
- PMID: 20618897
- PMCID: PMC3060346
- DOI: 10.1111/j.1365-294X.2010.04679.x
The landscape genetics of infectious disease emergence and spread
Abstract
The spread of parasites is inherently a spatial process often embedded in physically complex landscapes. It is therefore not surprising that infectious disease researchers are increasingly taking a landscape genetics perspective to elucidate mechanisms underlying basic ecological processes driving infectious disease dynamics and to understand the linkage between spatially dependent population processes and the geographic distribution of genetic variation within both hosts and parasites. The increasing availability of genetic information on hosts and parasites when coupled to their ecological interactions can lead to insights for predicting patterns of disease emergence, spread and control. Here, we review research progress in this area based on four different motivations for the application of landscape genetics approaches: (i) assessing the spatial organization of genetic variation in parasites as a function of environmental variability, (ii) using host population genetic structure as a means to parameterize ecological dynamics that indirectly influence parasite populations, for example, gene flow and movement pathways across heterogeneous landscapes and the concurrent transport of infectious agents, (iii) elucidating the temporal and spatial scales of disease processes and (iv) reconstructing and understanding infectious disease invasion. Throughout this review, we emphasize that landscape genetic principles are relevant to infection dynamics across a range of scales from within host dynamics to global geographic patterns and that they can also be applied to unconventional 'landscapes' such as heterogeneous contact networks underlying the spread of human and livestock diseases. We conclude by discussing some general considerations and problems for inferring epidemiological processes from genetic data and try to identify possible future directions and applications for this rapidly expanding field.
Figures
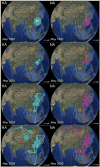
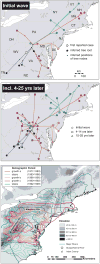
References
-
- Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends in Ecology & Evolution. 2009;24:21–30. - PubMed
-
- Balkenhol N, Gugerli F, Cushman SA, et al. Identifying future research needs in landscape genetics: where to from here? Landscape Ecology. 2009;24:455–463.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

