HIV-1 Nef binds a subpopulation of MHC-I throughout its trafficking itinerary and down-regulates MHC-I by perturbing both anterograde and retrograde trafficking
- PMID: 20622010
- PMCID: PMC2945581
- DOI: 10.1074/jbc.M110.135947
HIV-1 Nef binds a subpopulation of MHC-I throughout its trafficking itinerary and down-regulates MHC-I by perturbing both anterograde and retrograde trafficking
Erratum in
- J Biol Chem. 2010 Dec 3;285(49):38740. Chaudhury, Bhabhadeb [corrected to Chowdhury, Bhabadeb]
Abstract
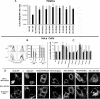
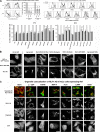
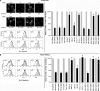
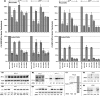
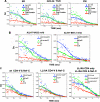
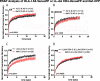
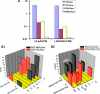
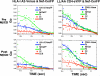
The HIV protein Nef is thought to mediate immune evasion and promote viral persistence in part by down-regulating major histocompatibility complex class I protein (MHC-I or HLA-I) from the cell surface. Two different models have been proposed to explain this phenomenon as follows: 1) stimulation of MHC-I retrograde trafficking from and aberrant recycling to the plasma membrane, and 2) inhibition of anterograde trafficking of newly synthesized HLA-I from the endoplasmic reticulum to the plasma membrane. We show here that Nef simultaneously uses both mechanisms to down-regulate HLA-I in peripheral blood mononuclear cells or HeLa cells. Consistent with this, we found by using fluorescence correlation spectroscopy that a third of diffusing HLA-I at the endoplasmic reticulum, Golgi/trans-Golgi network, and the plasma membrane (PM) was associated with Nef. The binding of Nef was similarly avid for native HLA-I and recombinant HLA-I A2 at the PM. Nef binding to HLA-I at the PM was sensitive to specific inhibition of endocytosis. It was also attenuated by cyclodextrin disruption of PM lipid micro-domain architecture, a change that also retarded lateral diffusion and induced large clusters of HLA-I. In all, our data support a model for Nef down-regulation of HLA-I that involves both major trafficking itineraries and persistent protein-protein interactions throughout the cell.
Figures

Similar articles
-
The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes.EMBO J. 1998 May 15;17(10):2777-89. doi: 10.1093/emboj/17.10.2777. EMBO J. 1998. PMID: 9582271 Free PMC article.
-
HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling.Sci Rep. 2016 Nov 14;6:37021. doi: 10.1038/srep37021. Sci Rep. 2016. PMID: 27841315 Free PMC article.
-
Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking.J Virol. 2002 Dec;76(23):12173-84. doi: 10.1128/jvi.76.23.12173-12184.2002. J Virol. 2002. PMID: 12414957 Free PMC article.
-
HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion.Biochim Biophys Acta. 2015 Apr;1850(4):733-41. doi: 10.1016/j.bbagen.2015.01.003. Epub 2015 Jan 10. Biochim Biophys Acta. 2015. PMID: 25585010 Review.
-
Human leukocyte antigen (HLA) class I down-regulation by human immunodeficiency virus type 1 negative factor (HIV-1 Nef): what might we learn from natural sequence variants?Viruses. 2012 Sep;4(9):1711-30. doi: 10.3390/v4091711. Epub 2012 Sep 24. Viruses. 2012. PMID: 23170180 Free PMC article. Review.
Cited by
-
ADP ribosylation factor 1 activity is required to recruit AP-1 to the major histocompatibility complex class I (MHC-I) cytoplasmic tail and disrupt MHC-I trafficking in HIV-1-infected primary T cells.J Virol. 2011 Dec;85(23):12216-26. doi: 10.1128/JVI.00056-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917951 Free PMC article.
-
Brain transcriptome-wide screen for HIV-1 Nef protein interaction partners reveals various membrane-associated proteins.PLoS One. 2012;7(12):e51578. doi: 10.1371/journal.pone.0051578. Epub 2012 Dec 17. PLoS One. 2012. PMID: 23284715 Free PMC article.
-
Coronavirus Porcine Deltacoronavirus Upregulates MHC Class I Expression through RIG-I/IRF1-Mediated NLRC5 Induction.J Virol. 2022 Apr 13;96(7):e0015822. doi: 10.1128/jvi.00158-22. Epub 2022 Mar 21. J Virol. 2022. PMID: 35311551 Free PMC article.
-
The Small GTPase Arf6: An Overview of Its Mechanisms of Action and of Its Role in Host⁻Pathogen Interactions and Innate Immunity.Int J Mol Sci. 2019 May 5;20(9):2209. doi: 10.3390/ijms20092209. Int J Mol Sci. 2019. PMID: 31060328 Free PMC article. Review.
-
HIV-1 Nef disrupts intracellular trafficking of major histocompatibility complex class I, CD4, CD8, and CD28 by distinct pathways that share common elements.J Virol. 2011 Jul;85(14):6867-81. doi: 10.1128/JVI.00229-11. Epub 2011 May 4. J Virol. 2011. PMID: 21543478 Free PMC article.
References
-
- Collins K. L., Baltimore D. (1999) Immunol. Rev. 168, 65–74 - PubMed
-
- Garcia J. V., Miller A. D. (1991) Nature 350, 508–511 - PubMed
-
- Schwartz O., Maréchal V., Le Gall S., Lemonnier F., Heard J. M. (1996) Nat. Med. 2, 338–342 - PubMed
-
- Sol-Foulon N., Moris A., Nobile C., Boccaccio C., Engering A., Abastado J. P., Heard J. M., van Kooyk Y., Schwartz O. (2002) Immunity 16, 145–155 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

