Superinhibitory phospholamban mutants compete with Ca2+ for binding to SERCA2a by stabilizing a unique nucleotide-dependent conformational state
- PMID: 20622261
- PMCID: PMC2937880
- DOI: 10.1074/jbc.M110.151779
Superinhibitory phospholamban mutants compete with Ca2+ for binding to SERCA2a by stabilizing a unique nucleotide-dependent conformational state
Abstract
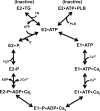
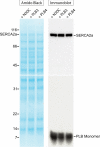
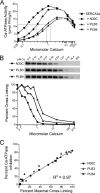
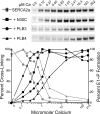
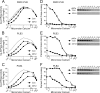
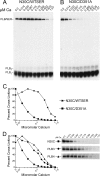
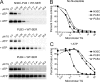
Three cross-linkable phospholamban (PLB) mutants of increasing inhibitory strength (N30C-PLB < N27A,N30C,L37A-PLB (PLB3) < N27A,N30C,L37A,V49G-PLB (PLB4)) were used to determine whether PLB decreases the Ca(2+) affinity of SERCA2a by competing for Ca(2+) binding. The functional effects of N30C-PLB, PLB3, and PLB4 on Ca(2+)-ATPase activity and E1 approximately P formation were correlated with their binding interactions with SERCA2a measured by chemical cross-linking. Successively higher Ca(2+) concentrations were required to both activate the enzyme co-expressed with N30C-PLB, PLB3, and PLB4 and to dissociate N30C-PLB, PLB3, and PLB4 from SERCA2a, suggesting competition between PLB and Ca(2+) for binding to SERCA2a. This was confirmed with the Ca(2+) pump mutant, D351A, which is catalytically inactive but retains strong Ca(2+) binding. Increasingly higher Ca(2+) concentrations were also required to dissociate N30C-PLB, PLB3, and PLB4 from D351A, demonstrating directly that PLB antagonizes Ca(2+) binding. Finally, the specific conformation of E2 (Ca(2+)-free state of SERCA2a) that binds PLB was investigated using the Ca(2+)-pump inhibitors thapsigargin and vanadate. Cross-linking assays conducted in the absence of Ca(2+) showed that PLB bound preferentially to E2 with bound nucleotide, forming a remarkably stable complex that is highly resistant to both thapsigargin and vanadate. In the presence of ATP, N30C-PLB had an affinity for SERCA2a approaching that of vanadate (micromolar), whereas PLB3 and PLB4 had much higher affinities, severalfold greater than even thapsigargin (nanomolar or higher). We conclude that PLB decreases Ca(2+) binding to SERCA2a by stabilizing a unique E2.ATP state that is unable to bind thapsigargin or vanadate.
Figures

References
-
- Simmerman H. K., Jones L. R. (1998) Physiol. Rev. 78, 921–947 - PubMed
-
- MacLennan D. H., Kranias E. G. (2003) Nat. Rev. Mol. Cell Biol. 4, 566–577 - PubMed
-
- Kimura Y., Kurzydlowski K., Tada M., MacLennan D. H. (1997) J. Biol. Chem. 272, 15061–16064 - PubMed
-
- Autry J. M., Jones L. R. (1997) J. Biol. Chem. 272, 15872–15880 - PubMed
-
- Cornea R. L., Jones L. R., Autry J. M., Thomas D. D. (1997) Biochemistry 36, 2960–2967 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

