Comparison of 61 sequenced Escherichia coli genomes
- PMID: 20623278
- PMCID: PMC2974192
- DOI: 10.1007/s00248-010-9717-3
Comparison of 61 sequenced Escherichia coli genomes
Abstract
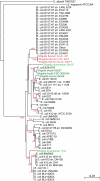
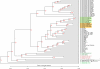
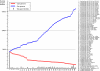
Escherichia coli is an important component of the biosphere and is an ideal model for studies of processes involved in bacterial genome evolution. Sixty-one publically available E. coli and Shigella spp. sequenced genomes are compared, using basic methods to produce phylogenetic and proteomics trees, and to identify the pan- and core genomes of this set of sequenced strains. A hierarchical clustering of variable genes allowed clear separation of the strains into clusters, including known pathotypes; clinically relevant serotypes can also be resolved in this way. In contrast, when in silico MLST was performed, many of the various strains appear jumbled and less well resolved. The predicted pan-genome comprises 15,741 gene families, and only 993 (6%) of the families are represented in every genome, comprising the core genome. The variable or 'accessory' genes thus make up more than 90% of the pan-genome and about 80% of a typical genome; some of these variable genes tend to be co-localized on genomic islands. The diversity within the species E. coli, and the overlap in gene content between this and related species, suggests a continuum rather than sharp species borders in this group of Enterobacteriaceae.
Figures



References
-
- Blattner FR, Plunkett G3, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. - DOI - PubMed
-
- Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Genomic Standards Consortium Human Microbiome Project Jumpstart Consortium. Detter JC. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. - DOI - PMC - PubMed
-
- Chen SL, Hung C, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sc USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

