Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes
- PMID: 20624288
- PMCID: PMC2926783
- DOI: 10.1186/gb-2010-11-7-r72
Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes
Abstract
Background: Long considered to be the building block of life, it is now apparent that protein is only one of many functional products generated by the eukaryotic genome. Indeed, more of the human genome is transcribed into noncoding sequence than into protein-coding sequence. Nevertheless, whilst we have developed a deep understanding of the relationships between evolutionary constraint and function for protein-coding sequence, little is known about these relationships for non-coding transcribed sequence. This dearth of information is partially attributable to a lack of established non-protein-coding RNA (ncRNA) orthologs among birds and mammals within sequence and expression databases.
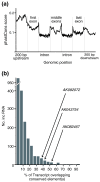
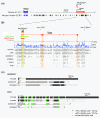
Results: Here, we performed a multi-disciplinary study of four highly conserved and brain-expressed transcripts selected from a list of mouse long intergenic noncoding RNA (lncRNA) loci that generally show pronounced evolutionary constraint within their putative promoter regions and across exon-intron boundaries. We identify some of the first lncRNA orthologs present in birds (chicken), marsupial (opossum), and eutherian mammals (mouse), and investigate whether they exhibit conservation of brain expression. In contrast to conventional protein-coding genes, the sequences, transcriptional start sites, exon structures, and lengths for these non-coding genes are all highly variable.
Conclusions: The biological relevance of lncRNAs would be highly questionable if they were limited to closely related phyla. Instead, their preservation across diverse amniotes, their apparent conservation in exon structure, and similarities in their pattern of brain expression during embryonic and early postnatal stages together indicate that these are functional RNA molecules, of which some have roles in vertebrate brain development.
Figures




References
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
-
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, Hlavina W, Kapustin Y, Meric P, Maglott D, Birtle Z, Marques AC, Graves T, Zhou S, Teague B, Potamousis K, Churas C, Place M, Herschleb J, Runnheim R, Forrest D, Amos-Landgraf J, Schwartz DC, Cheng Z, Lindblad-Toh K, Eichler EE. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. - DOI - PMC - PubMed
-
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Baily DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/F007590/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U137761446/MRC_/Medical Research Council/United Kingdom
- MC_U137761449/MRC_/Medical Research Council/United Kingdom
- BB/F003285/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

