Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda
- PMID: 20624745
- PMCID: PMC2998192
- DOI: 10.1093/gbe/evq030
Ecdysozoan mitogenomics: evidence for a common origin of the legged invertebrates, the Panarthropoda
Abstract
Ecdysozoa is the recently recognized clade of molting animals that comprises the vast majority of extant animal species and the most important invertebrate model organisms--the fruit fly and the nematode worm. Evolutionary relationships within the ecdysozoans remain, however, unresolved, impairing the correct interpretation of comparative genomic studies. In particular, the affinities of the three Panarthropoda phyla (Arthropoda, Onychophora, and Tardigrada) and the position of Myriapoda within Arthropoda (Mandibulata vs. Myriochelata hypothesis) are among the most contentious issues in animal phylogenetics. To elucidate these relationships, we have determined and analyzed complete or nearly complete mitochondrial genome sequences of two Tardigrada, Hypsibius dujardini and Thulinia sp. (the first genomes to date for this phylum); one Priapulida, Halicryptus spinulosus; and two Onychophora, Peripatoides sp. and Epiperipatus biolleyi; and a partial mitochondrial genome sequence of the Onychophora Euperipatoides kanagrensis. Tardigrada mitochondrial genomes resemble those of the arthropods in term of the gene order and strand asymmetry, whereas Onychophora genomes are characterized by numerous gene order rearrangements and strand asymmetry variations. In addition, Onychophora genomes are extremely enriched in A and T nucleotides, whereas Priapulida and Tardigrada are more balanced. Phylogenetic analyses based on concatenated amino acid coding sequences support a monophyletic origin of the Ecdysozoa and the position of Priapulida as the sister group of a monophyletic Panarthropoda (Tardigrada plus Onychophora plus Arthropoda). The position of Tardigrada is more problematic, most likely because of long branch attraction (LBA). However, experiments designed to reduce LBA suggest that the most likely placement of Tardigrada is as a sister group of Onychophora. The same analyses also recover monophyly of traditionally recognized arthropod lineages such as Arachnida and of the highly debated clade Mandibulata.
Figures

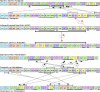
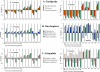
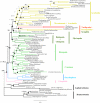



References
-
- Abascal F, Posada D, Zardoya R. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 2007;24:1–5. - PubMed
-
- Adachi J, Hasegawa M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 1996;42:459–468. - PubMed
-
- Aguinaldo AM, et al. Evidence for a clade of nematodes, arthropods, and other moulting animals. Nature. 1997;387:489–493. - PubMed
-
- Blanquart S, Lartillot N. A site- and time-heterogeneous model of amino acid replacement. Mol Biol Evol. 2008;25:842–858. - PubMed
-
- Boore JL, Lavrov DV, Brown WM. Gene translocation links insects and crustaceans. Nature. 1998;392:667–668. - PubMed

